Index >
Results for CNB 83240-rev
Input Alignment
CLUSTAL W(1.81) multiple sequence alignment
83240_ENSG00000075292_HUMAN_9792_9941/1-155 GCAACGGCATTCAGCTGCTGAAGCGCCAACTGAGTCTTAATCTGAGCAAAGTGCAAAGGA
83240_ENSDARG00000004048_ZEBRAFISH_8782_8931/1-155 -CAATTGCATTCAGCTGGTGCAGTGCCAACTGAGTCTTTATCTGTGCCAACTGC-AGGGA
83240_ENSMUSG00000030016_MOUSE_9792_9941/1-155 GCAACGGCATTCAGCTGCTGAAGCGCCAACTGAGTCTCAATCTGAGCAAAGCGCAAAGGA
83240_ENSRNOG00000014501_RAT_9792_9941/1-155 GCAACGGCATTCAGCTGCTGAAGCGCCAACTGAGTCTTAATCTGAGCAAAGTGCAAAGGA
*** *********** ** ** ************* ***** ** ** ** * ***
83240_ENSG00000075292_HUMAN_9792_9941/1-155 GA-TGGGCCCAACAGAAGTGGGTTTGCAATGCCCAGGTTCAAGGAATT---CACAAGTGG
83240_ENSDARG00000004048_ZEBRAFISH_8782_8931/1-155 GACAGGGCTTAAGATGGGCAGATTCCCAA---AATGTCTCACGGAATTTGGAACAGGGTG
83240_ENSMUSG00000030016_MOUSE_9792_9941/1-155 GA-CGGGCCCAACAGAAGTGGGCTCGCAATGCCCAGGCTCAAGGAATT---CACAAGTGG
83240_ENSRNOG00000014501_RAT_9792_9941/1-155 GA-TGGGCCCAACAGAAGTGGGTTTGCAATGCCCAGGCTCAAGGAATT---CACAAGTGG
** **** ** * * * * *** * *** ****** *** * *
83240_ENSG00000075292_HUMAN_9792_9941/1-155 TGCGAAATTTTCCAAGAATAACACAAAGCTGAAA-
83240_ENSDARG00000004048_ZEBRAFISH_8782_8931/1-155 CACGCAACTTTCCAAGAACAACGAAAAGCTGGAAA
83240_ENSMUSG00000030016_MOUSE_9792_9941/1-155 TGCGAAATTTTCCAAGAATAACACAAAGCTGAAA-
83240_ENSRNOG00000014501_RAT_9792_9941/1-155 TGCGAAATTTTCCAAGAATAACACAAAGCTGAAA-
** ** ********** *** ******* **
83240-rev.aln
RNAz output
############################ RNAz 0.1 ##############################
Sequences: 4
Columns: 155
Mean pairwise identity: 82.30
Mean single sequence MFE: -44.13
Consensus MFE: -28.98
Energy contribution: -28.10
Covariance contribution: -0.87
Mean z-score: -1.67
Structure conservation index: 0.66
SVM decision value: 0.10
SVM RNA-class probability: 0.582765
Prediction: RNA
######################################################################
>83240_ENSG00000075292_HUMAN_9792_9941/1-155
GCAACGGCAUUCAGCUGCUGAAGCGCCAACUGAGUCUUAAUCUGAGCAAAGUGCAAAGGAGAUGGGCCCAACAGAAGUGGGUUUGCAAUGCCCAGGUUCAAGGAAUUCACAAGUGGUGCGAAAUUUUCCAAGAAUAACACAAAGCUGAAA
((....)).(((((((..((..((((((..((((((((((((((.(((...(((((.........(((((.......)))))))))).))).))))))..))).)))))....))))))...((((.....))))....)).))))))). ( -44.10)
>83240_ENSDARG00000004048_ZEBRAFISH_8782_8931/1-155
CAAUUGCAUUCAGCUGGUGCAGUGCCAACUGAGUCUUUAUCUGUGCCAACUGCAGGGAGACAGGGCUUAAGAUGGGCAGAUUCCCAAAAUGUCUCACGGAAUUUGGAACAGGGUGCACGCAACUUUCCAAGAACAACGAAAAGCUGGAAA
........(((((((.(((((.(.(((.(((((((((..(((.(((.....))).)))...))))))).)).))).((((((((.............))))))))......).))))).....((((..........))))))))))).. ( -41.42)
>83240_ENSMUSG00000030016_MOUSE_9792_9941/1-155
GCAACGGCAUUCAGCUGCUGAAGCGCCAACUGAGUCUCAAUCUGAGCAAAGCGCAAAGGAGACGGGCCCAACAGAAGUGGGCUCGCAAUGCCCAGGCUCAAGGAAUUCACAAGUGGUGCGAAAUUUUCCAAGAAUAACACAAAGCUGAAA
((....)).(((((((..((..((((((..(((((..(....(((((...(.(((..(....)(((((((.......)))))))....))))...))))).)..)))))....))))))...((((.....))))....)).))))))). ( -49.10)
>83240_ENSRNOG00000014501_RAT_9792_9941/1-155
GCAACGGCAUUCAGCUGCUGAAGCGCCAACUGAGUCUUAAUCUGAGCAAAGUGCAAAGGAGAUGGGCCCAACAGAAGUGGGUUUGCAAUGCCCAGGCUCAAGGAAUUCACAAGUGGUGCGAAAUUUUCCAAGAAUAACACAAAGCUGAAA
((....)).(((((((..((..((((((..((((((((..(((..((.....))..)))...((((((.........(((((.......)))))))))))))).)))))....))))))...((((.....))))....)).))))))). ( -41.90)
>consensus
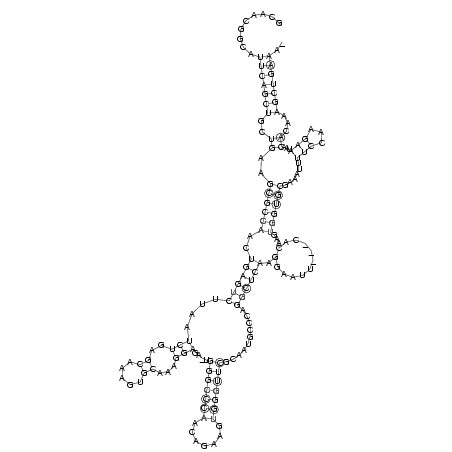
GCAACGGCAUUCAGCUGCUGAAGCGCCAACUGAGUCUUAAUCUGAGCAAAGUGCAAAGGAGA_UGGGCCCAACAGAAGUGGGUUCGCAAUGCCCAGGCUCAAGGAAUU___CACAAGUGGUGCGAAAUUUUCCAAGAAUAACACAAAGCUGAAA_
.........(((((((..((..((((((..((((((....(((..((.....))...)))....(((((((.......)))))))..........)))))).(..........)...))))))......(((...)))...))...))))))).. (-28.98 = -28.10 + -0.87)
83240-rev.rnaz
RNAalifold consensus structure
Stefan Washietl
Last modified: Thu Aug 12 14:35:44 CEST 2004