Index >
Results for CNB 502451-rev
Input Alignment
CLUSTAL W(1.81) multiple sequence alignment
502451_ENSG00000166535_HUMAN_9684_9739/1-58 -TAGATGATGGGAGCAAACAAGACCATGTTCTGCTCGCCACAGCCACTGGGCATCTG-
502451_ENSMUSG00000052587_MOUSE_1864_1921/1-58 GTAGATGTTGGGAGCAAAAAGGACCATGTTCTGCTCCCCACAGCCATAGGGCATTTGG
502451_ENSRNOG00000014814_RAT_8495_8552/1-58 ATAGATGTTTGGAGCAAAAAGGACCATGTTCTGCTCCCCACAGCCATAGGGCATTTGG
502451_ENSDARG00000008835_ZEBRAFISH_2351_2408/1-58 GTAAATATTGGGAGCGAGAATGATCATGTTCTGTTCTCCACAGCCATAAGGCATCTGT
** ** * ***** * * ** ********* ** ********* ***** **
502451-rev.aln
RNAz output
############################ RNAz 0.1 ##############################
Sequences: 4
Columns: 58
Mean pairwise identity: 82.76
Mean single sequence MFE: -17.50
Consensus MFE: -12.11
Energy contribution: -12.93
Covariance contribution: 0.81
Mean z-score: -1.81
Structure conservation index: 0.69
SVM decision value: 0.45
SVM RNA-class probability: 0.739415
Prediction: RNA
######################################################################
>502451_ENSG00000166535_HUMAN_9684_9739/1-58
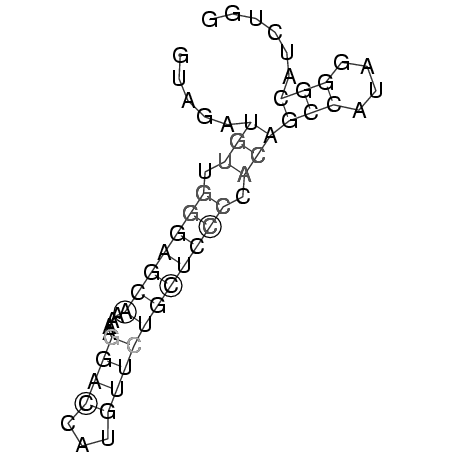
UAGAUGAUGGGAGCAAACAAGACCAUGUUCUGCUCGCCACAGCCACUGGGCAUCUG
(((((..((((((((((((......)))).))))).)))..(((....)))))))) ( -18.80)
>502451_ENSMUSG00000052587_MOUSE_1864_1921/1-58
GUAGAUGUUGGGAGCAAAAAGGACCAUGUUCUGCUCCCCACAGCCAUAGGGCAUUUGG
.....(((.(((((((....((((...))))))))))).)))(((....)))...... ( -20.00)
>502451_ENSRNOG00000014814_RAT_8495_8552/1-58
AUAGAUGUUUGGAGCAAAAAGGACCAUGUUCUGCUCCCCACAGCCAUAGGGCAUUUGG
.....(((..((((((....((((...))))))))))..)))(((....)))...... ( -15.70)
>502451_ENSDARG00000008835_ZEBRAFISH_2351_2408/1-58
GUAAAUAUUGGGAGCGAGAAUGAUCAUGUUCUGUUCUCCACAGCCAUAAGGCAUCUGU
........(((((((.(((((......))))))))).)))..(((....)))...... ( -15.50)
>consensus
GUAGAUGUUGGGAGCAAAAAGGACCAUGUUCUGCUCCCCACAGCCAUAGGGCAUCUGG
.....(((.(((((((....((((...))))))))))).)))(((....)))...... (-12.11 = -12.93 + 0.81)
502451-rev.rnaz
RNAalifold consensus structure
Stefan Washietl
Last modified: Thu Aug 12 14:35:44 CEST 2004