Index >
Results for CNB 481656_3-rev
Input Alignment
CLUSTAL W(1.81) multiple sequence alignment
481656_ENSRNOG00000007049_RAT_9511_9952/1-454 CTCGGTTCACATCGGGAGAGCCGGGTTAGAAAGAAGGAGACTCCAGAGAAAATATCTTCA
481656_ENSG00000119866_HUMAN_9496_9947/1-454 CTCGGTTCACATCGGGAGAGCCGGGTTAGAAAGAAGGAGACTCCAGAGAAAATATCTTCA
481656_ENSMUSG00000000861_MOUSE_2861_3289/1-454 CTCGGTTCACATCGGGAGAGCCGGGTTAGAAAGAAGGAGACTCCAGAGAAAATATCTTCA
481656_ENSDARG00000018362_ZEBRAFISH_9467_9847/1-454 CTCGGTTCACATCGGGAGAGCCGGGTTAGA---AAGGAGACTCCAGAGAAAATATCTTCA
****************************** ***************************
481656_ENSRNOG00000007049_RAT_9511_9952/1-454 TCAGTGCCTTTTGACATCCAAAATAAATTAGAAATAATACAAAGATGGCGCAGGGAAGAT
481656_ENSG00000119866_HUMAN_9496_9947/1-454 TCAGTGCCTTTTGACATCCAAAATAAATTAGAAATAATACAAAGATGGCGCAGGGAAGAT
481656_ENSMUSG00000000861_MOUSE_2861_3289/1-454 TCAGTGCCTTTTGACATCCAAAATAAATTAGAAATAATACAAAGATGGCGCAGGGAAGAT
481656_ENSDARG00000018362_ZEBRAFISH_9467_9847/1-454 TCAATGCC-TTTGACATCCAAAATAAATTAGAAATAATACAAAGATGGCGCAGGGAAGAT
*** **** ***************************************************
481656_ENSRNOG00000007049_RAT_9511_9952/1-454 GAATTGTGGGAGAGCCGTCATGGC----------NNNNNNNNNNNNNNAAGAAAAAAAAA
481656_ENSG00000119866_HUMAN_9496_9947/1-454 GAATTGTGGGAGAGCCGTCATGGCTTTTTTTTAAGCNNNNNNNNNNNNNNNNNNNNNNNN
481656_ENSMUSG00000000861_MOUSE_2861_3289/1-454 GAATTGTGGGAGAGCCGTCATGGC---------------NNNNNNNNNNNNAAGGAAAAA
481656_ENSDARG00000018362_ZEBRAFISH_9467_9847/1-454 GGATTGTGGGATAGCCGTCATGGC---------------TTTTTTGAAAAGAGAGAGAGA
* ********* ************
481656_ENSRNOG00000007049_RAT_9511_9952/1-454 NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNTGAAAAAA
481656_ENSG00000119866_HUMAN_9496_9947/1-454 NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNTGAAAAAA
481656_ENSMUSG00000000861_MOUSE_2861_3289/1-454 ------AANNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNTGAAAAAA
481656_ENSDARG00000018362_ZEBRAFISH_9467_9847/1-454 ------------------------------------------------GAGATG-AAAAA
** *****
481656_ENSRNOG00000007049_RAT_9511_9952/1-454 ATGGCAAAAGCCCCCCTGAGCTGCAAGTTCAAGTGCGGACGTGACGTCCCTGCGAACTTG
481656_ENSG00000119866_HUMAN_9496_9947/1-454 ATGGCAAAAGCCCCCCTGAGCTGCAAGTTCAAGTGCGGACGTGACGTCCCTGCGAACTTG
481656_ENSMUSG00000000861_MOUSE_2861_3289/1-454 ATGGCAAAAGCCCCCCTGAGCTGCAAGTTCAAGTGCGGACGTGACGTCCCTGCGAACTTG
481656_ENSDARG00000018362_ZEBRAFISH_9467_9847/1-454 ATGGCAAAAGCCCCCCTGAGCTGCAAGTTCAAGTGCGGACGTGACGTCCCTGCGAACTTG
************************************************************
481656_3-rev.aln
RNAz output
############################ RNAz 0.1 ##############################
Sequences: 4
Columns: 300
Mean pairwise identity: 81.92
Mean single sequence MFE: -62.20
Consensus MFE: -54.78
Energy contribution: -57.02
Covariance contribution: 2.25
Mean z-score: -1.28
Structure conservation index: 0.88
SVM decision value: -0.01
SVM RNA-class probability: 0.528877
Prediction: RNA
######################################################################
>481656_ENSRNOG00000007049_RAT_9511_9952/1-454
CUCGGUUCACAUCGGGAGAGCCGGGUUAGAAAGAAGGAGACUCCAGAGAAAAUAUCUUCAUCAGUGCCUUUUGACAUCCAAAAUAAAUUAGAAAUAAUACAAAGAUGGCGCAGGGAAGAUGAAUUGUGGGAGAGCCGUCAUGGCNNNNNNNNNNNNNNAAGAAAAAAAAANNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNUGAAAAAAAUGGCAAAAGCCCCCCUGAGCUGCAAGUUCAAGUGCGGACGUGACGUCCCUGCGAACUUG
((((((((.(.....).)))))))).......(.((....)).).(((....(((((((.((.((((((((((.....)))))...((((....))))........))))).)))))))))..(((..((.((..(((((((..........................................................................................(((....)))(((((((((.....)))).)).).)).)))))))))))..))).))). ( -60.80)
>481656_ENSG00000119866_HUMAN_9496_9947/1-454
CUCGGUUCACAUCGGGAGAGCCGGGUUAGAAAGAAGGAGACUCCAGAGAAAAUAUCUUCAUCAGUGCCUUUUGACAUCCAAAAUAAAUUAGAAAUAAUACAAAGAUGGCGCAGGGAAGAUGAAUUGUGGGAGAGCCGUCAUGGCUUUUUUUUAAGCNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNUGAAAAAAAUGGCAAAAGCCCCCCUGAGCUGCAAGUUCAAGUGCGGACGUGACGUCCCUGCGAACUUG
((((((....))))))..(((((((((((((((((.....((((...((...(((((((.((.((((((((((.....)))))...((((....))))........))))).)))))))))..))...)))).(((.....)))))))))))))........................................................................................(((....)))..)))).))).(((((((..(((.(((((...))))).)))))))))) ( -63.40)
>481656_ENSMUSG00000000861_MOUSE_2861_3289/1-454
CUCGGUUCACAUCGGGAGAGCCGGGUUAGAAAGAAGGAGACUCCAGAGAAAAUAUCUUCAUCAGUGCCUUUUGACAUCCAAAAUAAAUUAGAAAUAAUACAAAGAUGGCGCAGGGAAGAUGAAUUGUGGGAGAGCCGUCAUGGCNNNNNNNNNNNNAAGGAAAAAAANNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNUGAAAAAAAUGGCAAAAGCCCCCCUGAGCUGCAAGUUCAAGUGCGGACGUGACGUCCCUGCGAACUUG
((((((((.(.....).)))))))).......(.((....)).).(((....(((((((.((.((((((((((.....)))))...((((....))))........))))).)))))))))..(((..((.((..(((((((...............................................................................(((....)))(((((((((.....)))).)).).)).)))))))))))..))).))). ( -60.80)
>481656_ENSDARG00000018362_ZEBRAFISH_9467_9847/1-454
CUCGGUUCACAUCGGGAGAGCCGGGUUAGAAAGGAGACUCCAGAGAAAAUAUCUUCAUCAAUGCCUUUGACAUCCAAAAUAAAUUAGAAAUAAUACAAAGAUGGCGCAGGGAAGAUGGAUUGUGGGAUAGCCGUCAUGGCUUUUUUGAAAAGAGAGAGAGAGAGAUGAAAAAAUGGCAAAAGCCCCCCUGAGCUGCAAGUUCAAGUGCGGACGUGACGUCCCUGCGAACUUG
..(((((((....(((...(((((((((...((....))(((.((....(((((((.((..((((((((.....))))....((((....))))........))))..)))))))))..)).)))..))))).((((..(((((((.........)))))))..)))).....)))).....)))...)))))))(((((((..(((.(((((...))))).)))))))))) ( -63.80)
>consensus
CUCGGUUCACAUCGGGAGAGCCGGGUUAGAAAGAAGGAGACUCCAGAGAAAAUAUCUUCAUCAGUGCCUUUUGACAUCCAAAAUAAAUUAGAAAUAAUACAAAGAUGGCGCAGGGAAGAUGAAUUGUGGGAGAGCCGUCAUGGC___________________________AAAGAAAAA____________________________________________________UGAAAAAAAUGGCAAAAGCCCCCCUGAGCUGCAAGUUCAAGUGCGGACGUGACGUCCCUGCGAACUUG
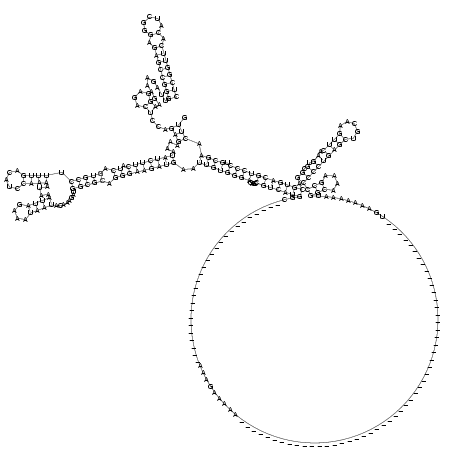
((((((((.(.....).)))))))).........((....))...(((....(((((((.((.(((((.((((.....))))....((((....))))........))))).)))))))))..((((((((....(((((((....................................................................................................(((....)))(((((((((.....)))).)).).)).))))))))))).)))).))). (-54.78 = -57.02 + 2.25)
481656_3-rev.rnaz
RNAalifold consensus structure
Stefan Washietl
Last modified: Thu Aug 12 14:35:44 CEST 2004