Index >
Results for CNB 481656_2-rev
Input Alignment
CLUSTAL W(1.81) multiple sequence alignment
481656_ENSRNOG00000007049_RAT_9511_9952/1-454 AATATCTTCATCAGTGCCTTTTGACATCCAAAATAAATTAGAAATAATACAAAGATGGCG
481656_ENSG00000119866_HUMAN_9496_9947/1-454 AATATCTTCATCAGTGCCTTTTGACATCCAAAATAAATTAGAAATAATACAAAGATGGCG
481656_ENSMUSG00000000861_MOUSE_2861_3289/1-454 AATATCTTCATCAGTGCCTTTTGACATCCAAAATAAATTAGAAATAATACAAAGATGGCG
481656_ENSDARG00000018362_ZEBRAFISH_9467_9847/1-454 AATATCTTCATCAATGCC-TTTGACATCCAAAATAAATTAGAAATAATACAAAGATGGCG
************* **** *****************************************
481656_ENSRNOG00000007049_RAT_9511_9952/1-454 CAGGGAAGATGAATTGTGGGAGAGCCGTCATGGC----------NNNNNNNNNNNNNNAA
481656_ENSG00000119866_HUMAN_9496_9947/1-454 CAGGGAAGATGAATTGTGGGAGAGCCGTCATGGCTTTTTTTTAAGCNNNNNNNNNNNNNN
481656_ENSMUSG00000000861_MOUSE_2861_3289/1-454 CAGGGAAGATGAATTGTGGGAGAGCCGTCATGGC---------------NNNNNNNNNNN
481656_ENSDARG00000018362_ZEBRAFISH_9467_9847/1-454 CAGGGAAGATGGATTGTGGGATAGCCGTCATGGC---------------TTTTTTGAAAA
*********** ********* ************
481656_ENSRNOG00000007049_RAT_9511_9952/1-454 GAAAAAAAAANNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN
481656_ENSG00000119866_HUMAN_9496_9947/1-454 NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN
481656_ENSMUSG00000000861_MOUSE_2861_3289/1-454 NAAGGAAAAA------AANNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN
481656_ENSDARG00000018362_ZEBRAFISH_9467_9847/1-454 GAGAGAGAGA------------------------------------------------GA
481656_ENSRNOG00000007049_RAT_9511_9952/1-454 NNTGAAAAAAATGGCAAAAGCCCCCCTGAGCTGCAAGTTCAAGTGCGGACGTGACGTCCC
481656_ENSG00000119866_HUMAN_9496_9947/1-454 NNTGAAAAAAATGGCAAAAGCCCCCCTGAGCTGCAAGTTCAAGTGCGGACGTGACGTCCC
481656_ENSMUSG00000000861_MOUSE_2861_3289/1-454 NNTGAAAAAAATGGCAAAAGCCCCCCTGAGCTGCAAGTTCAAGTGCGGACGTGACGTCCC
481656_ENSDARG00000018362_ZEBRAFISH_9467_9847/1-454 GATG-AAAAAATGGCAAAAGCCCCCCTGAGCTGCAAGTTCAAGTGCGGACGTGACGTCCC
** *******************************************************
481656_ENSRNOG00000007049_RAT_9511_9952/1-454 TGCGAACTTGAACGTCAGGAGTCTGGATGGACAGAGACACACAAAACATGGGCAGGGC-G
481656_ENSG00000119866_HUMAN_9496_9947/1-454 TGCGAACTTGAACGTCAGGAGTCTGGATGGACAGAGACACACAAAACATGGGCAGGGC-G
481656_ENSMUSG00000000861_MOUSE_2861_3289/1-454 TGCGAACTTGAACGTCAGGAGTCTGGATGGACAGAGACACACAAAACATGGGCAGGGC-G
481656_ENSDARG00000018362_ZEBRAFISH_9467_9847/1-454 TGCGAACTTGAACGTCAGGAG-ATGGATGGACACAGACATACAGAACATGGGCAGGGCAA
********************* ********** ***** *** **************
481656_2-rev.aln
RNAz output
############################ RNAz 0.1 ##############################
Sequences: 4
Columns: 300
Mean pairwise identity: 81.21
Mean single sequence MFE: -62.80
Consensus MFE: -54.06
Energy contribution: -56.80
Covariance contribution: 2.75
Mean z-score: -1.82
Structure conservation index: 0.86
SVM decision value: 1.00
SVM RNA-class probability: 0.897705
Prediction: RNA
######################################################################
>481656_ENSRNOG00000007049_RAT_9511_9952/1-454
AAUAUCUUCAUCAGUGCCUUUUGACAUCCAAAAUAAAUUAGAAAUAAUACAAAGAUGGCGCAGGGAAGAUGAAUUGUGGGAGAGCCGUCAUGGCNNNNNNNNNNNNNNAAGAAAAAAAAANNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNUGAAAAAAAUGGCAAAAGCCCCCCUGAGCUGCAAGUUCAAGUGCGGACGUGACGUCCCUGCGAACUUGAACGUCAGGAGUCUGGAUGGACAGAGACACACAAAACAUGGGCAGGGCG
..(((((((.((.((((((((((.....)))))...((((....))))........))))).)))))))))............((((((((.........................................................................................)))))....((((.(((((....(((((((..(((.(((((...))))).))))))))))....)))))..((((......))))..............))))..))). ( -61.14)
>481656_ENSG00000119866_HUMAN_9496_9947/1-454
AAUAUCUUCAUCAGUGCCUUUUGACAUCCAAAAUAAAUUAGAAAUAAUACAAAGAUGGCGCAGGGAAGAUGAAUUGUGGGAGAGCCGUCAUGGCUUUUUUUUAAGCNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNUGAAAAAAAUGGCAAAAGCCCCCCUGAGCUGCAAGUUCAAGUGCGGACGUGACGUCCCUGCGAACUUGAACGUCAGGAGUCUGGAUGGACAGAGACACACAAAACAUGGGCAGGGCG
..(((((((.((.((((((((((.....)))))...((((....))))........))))).)))))))))..(((..((((((((.....))))))))..))).........................................................................................((....((((.(((((....(((((((..(((.(((((...))))).))))))))))....)))))..((((......))))..............))))...)). ( -65.10)
>481656_ENSMUSG00000000861_MOUSE_2861_3289/1-454
AAUAUCUUCAUCAGUGCCUUUUGACAUCCAAAAUAAAUUAGAAAUAAUACAAAGAUGGCGCAGGGAAGAUGAAUUGUGGGAGAGCCGUCAUGGCNNNNNNNNNNNNAAGGAAAAAAANNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNUGAAAAAAAUGGCAAAAGCCCCCCUGAGCUGCAAGUUCAAGUGCGGACGUGACGUCCCUGCGAACUUGAACGUCAGGAGUCUGGAUGGACAGAGACACACAAAACAUGGGCAGGGCG
..(((((((.((.((((((((((.....)))))...((((....))))........))))).)))))))))............((((((((..............................................................................)))))....((((.(((((....(((((((..(((.(((((...))))).))))))))))....)))))..((((......))))..............))))..))). ( -61.28)
>481656_ENSDARG00000018362_ZEBRAFISH_9467_9847/1-454
AAUAUCUUCAUCAAUGCCUUUGACAUCCAAAAUAAAUUAGAAAUAAUACAAAGAUGGCGCAGGGAAGAUGGAUUGUGGGAUAGCCGUCAUGGCUUUUUUGAAAAGAGAGAGAGAGAGAUGAAAAAAUGGCAAAAGCCCCCCUGAGCUGCAAGUUCAAGUGCGGACGUGACGUCCCUGCGAACUUGAACGUCAGGAGAUGGAUGGACACAGACAUACAGAACAUGGGCAGGGCAA
..(((((((.((..((((((((.....))))....((((....))))........))))..)))))))))............((((((((..(((((((.........)))))))..)))).....))))....(((((((((..(((...(((((((((((((((...)))))..))..))))))))........(((..((....))..))).)))..)).)))..)))).. ( -63.70)
>consensus
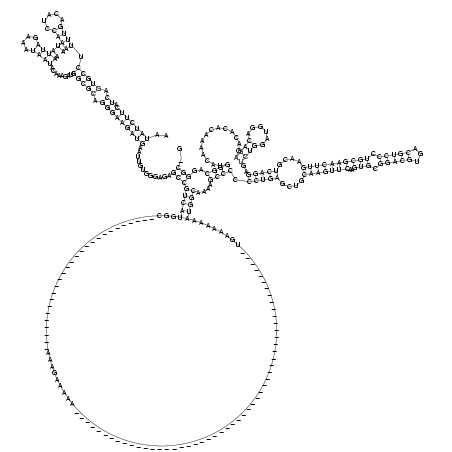
AAUAUCUUCAUCAGUGCCUUUUGACAUCCAAAAUAAAUUAGAAAUAAUACAAAGAUGGCGCAGGGAAGAUGAAUUGUGGGAGAGCCGUCAUGGC___________________________AAAGAAAAA____________________________________________________UGAAAAAAAUGGCAAAAGCCCCCCUGAGCUGCAAGUUCAAGUGCGGACGUGACGUCCCUGCGAACUUGAACGUCAGGAGUCUGGAUGGACAGAGACACACAAAACAUGGGCAGGGC_G
..(((((((.((.(((((.((((.....))))....((((....))))........))))).)))))))))............((((((((...................................................................................................)))))....((((.(((((....(((((((..(((.(((((...))))).))))))))))....)))))..((((......))))..............))))..))).. (-54.06 = -56.80 + 2.75)
481656_2-rev.rnaz
RNAalifold consensus structure
Stefan Washietl
Last modified: Thu Aug 12 14:35:44 CEST 2004