Index >
Results for CNB 481656_1
Input Alignment
CLUSTAL W(1.81) multiple sequence alignment
481656_ENSRNOG00000007049_RAT_9511_9952/1-454 GTGTCCATTGGTGT-GAGCTTCCCCTTCCCTCCTCCCCTTCTCTCCTGCTC-GCCCTGCC
481656_ENSG00000119866_HUMAN_9496_9947/1-454 GTGTCCATTGGTGT-GAGCTTCCCCTTCCCTCCTCCCCTTCTCTCCTGCTC-GCCCTGCC
481656_ENSMUSG00000000861_MOUSE_2861_3289/1-454 GTGTCCATTGGTGT-GAGCTTCCCCTTCCCTCCTCCCCTTCTCTCCTGCTC-GCCCTGCC
481656_ENSDARG00000018362_ZEBRAFISH_9467_9847/1-454 GTGTCCATTGGTTTTGTTATTTCCACTCCCTCCTCC--TACTCCACGGCTTTGCCCTGCC
481656_ENSRNOG00000007049_RAT_9511_9952/1-454 CATGTTTTGTGTGTCTCTGTCCATCCAGACTCCTGACGTTCAAGTTCGCAGGGACGTCAC
481656_ENSG00000119866_HUMAN_9496_9947/1-454 CATGTTTTGTGTGTCTCTGTCCATCCAGACTCCTGACGTTCAAGTTCGCAGGGACGTCAC
481656_ENSMUSG00000000861_MOUSE_2861_3289/1-454 CATGTTTTGTGTGTCTCTGTCCATCCAGACTCCTGACGTTCAAGTTCGCAGGGACGTCAC
481656_ENSDARG00000018362_ZEBRAFISH_9467_9847/1-454 CATGTTCTGTATGTCTGTGTCCATCCAT-CTCCTGACGTTCAAGTTCGCAGGGACGTCAC
481656_ENSRNOG00000007049_RAT_9511_9952/1-454 GTCCGCACTTGAACTTGCAGCTCAGGGGGGCTTTTGCCATTTTTTTCANNNNNNNNNNNN
481656_ENSG00000119866_HUMAN_9496_9947/1-454 GTCCGCACTTGAACTTGCAGCTCAGGGGGGCTTTTGCCATTTTTTTCANNNNNNNNNNNN
481656_ENSMUSG00000000861_MOUSE_2861_3289/1-454 GTCCGCACTTGAACTTGCAGCTCAGGGGGGCTTTTGCCATTTTTTTCANNNNNNNNNNNN
481656_ENSDARG00000018362_ZEBRAFISH_9467_9847/1-454 GTCCGCACTTGAACTTGCAGCTCAGGGGGGCTTTTGCCATTTTTT-CATCTC--------
481656_ENSRNOG00000007049_RAT_9511_9952/1-454 NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNTTTTTTTTTCTTNNNNNNNN
481656_ENSG00000119866_HUMAN_9496_9947/1-454 NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN
481656_ENSMUSG00000000861_MOUSE_2861_3289/1-454 NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNTT------TTTTTCCTTNNNNNNNNNNN
481656_ENSDARG00000018362_ZEBRAFISH_9467_9847/1-454 ----------------------------------------TCTCTCTCTCTTTTCAAAAA
481656_ENSRNOG00000007049_RAT_9511_9952/1-454 NNNNNN----------GCCATGACGGCTCTCCCACAATTCATCTTCCCTGCGCCATCTTT
481656_ENSG00000119866_HUMAN_9496_9947/1-454 NNNNGCTTAAAAAAAAGCCATGACGGCTCTCCCACAATTCATCTTCCCTGCGCCATCTTT
481656_ENSMUSG00000000861_MOUSE_2861_3289/1-454 N---------------GCCATGACGGCTCTCCCACAATTCATCTTCCCTGCGCCATCTTT
481656_ENSDARG00000018362_ZEBRAFISH_9467_9847/1-454 A---------------GCCATGACGGCTATCCCACAATCCATCTTCCCTGCGCCATCTTT
481656_1.aln
RNAz output
############################ RNAz 0.1 ##############################
Sequences: 4
Columns: 300
Mean pairwise identity: 79.12
Mean single sequence MFE: -57.34
Consensus MFE: -49.08
Energy contribution: -50.07
Covariance contribution: 1.00
Mean z-score: -2.86
Structure conservation index: 0.86
SVM decision value: 3.30
SVM RNA-class probability: 0.998954
Prediction: RNA
######################################################################
>481656_ENSRNOG00000007049_RAT_9511_9952/1-454
GUGUCCAUUGGUGUGAGCUUCCCCUUCCCUCCUCCCCUUCUCUCCUGCUCGCCCUGCCCAUGUUUUGUGUGUCUCUGUCCAUCCAGACUCCUGACGUUCAAGUUCGCAGGGACGUCACGUCCGCACUUGAACUUGCAGCUCAGGGGGGCUUUUGCCAUUUUUUUCANNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNUUUUUUUUUCUUNNNNNNNNNNNNNNGCCAUGACGGCUCUCCCACAAUUCAUCUUCCCUGCGCCAUCUUU
((((.....((.((((((............................)))))))).....(((..(((((.(...(((((........(((((((.(((((((((((..((((((...))))).)...)))))))).))))))))))(((....))).............................................................................................)))))....).)))))..))).......))))....... ( -57.99)
>481656_ENSG00000119866_HUMAN_9496_9947/1-454
GUGUCCAUUGGUGUGAGCUUCCCCUUCCCUCCUCCCCUUCUCUCCUGCUCGCCCUGCCCAUGUUUUGUGUGUCUCUGUCCAUCCAGACUCCUGACGUUCAAGUUCGCAGGGACGUCACGUCCGCACUUGAACUUGCAGCUCAGGGGGGCUUUUGCCAUUUUUUUCANNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNGCUUAAAAAAAAGCCAUGACGGCUCUCCCACAAUUCAUCUUCCCUGCGCCAUCUUU
((((.....((.((((((............................)))))))).....(((..(((((.(...(((((......(((((((((.(((((((((((..((((((...))))).)...)))))))).))))))))))(((....))).......)).............................................................................((((......))))...)))))....).)))))..))).......))))....... ( -60.19)
>481656_ENSMUSG00000000861_MOUSE_2861_3289/1-454
GUGUCCAUUGGUGUGAGCUUCCCCUUCCCUCCUCCCCUUCUCUCCUGCUCGCCCUGCCCAUGUUUUGUGUGUCUCUGUCCAUCCAGACUCCUGACGUUCAAGUUCGCAGGGACGUCACGUCCGCACUUGAACUUGCAGCUCAGGGGGGCUUUUGCCAUUUUUUUCANNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNUUUUUUUCCUUNNNNNNNNNNNNGCCAUGACGGCUCUCCCACAAUUCAUCUUCCCUGCGCCAUCUUU
((((.....((.((((((............................)))))))).....(((..(((((.(...(((((........(((((((.(((((((((((..((((((...))))).)...)))))))).))))))))))(((....)))..................................................................................)))))....).)))))..))).......))))....... ( -57.99)
>481656_ENSDARG00000018362_ZEBRAFISH_9467_9847/1-454
GUGUCCAUUGGUUUUGUUAUUUCCACUCCCUCCUCCUACUCCACGGCUUUGCCCUGCCCAUGUUCUGUAUGUCUGUGUCCAUCCAUCUCCUGACGUUCAAGUUCGCAGGGACGUCACGUCCGCACUUGAACUUGCAGCUCAGGGGGGCUUUUGCCAUUUUUUCAUCUCUCUCUCUCUCUUUUCAAAAAAGCCAUGACGGCUAUCCCACAAUCCAUCUUCCCUGCGCCAUCUUU
((((....(((..((((...........................(((...((((..(((.((..(((((.(((.(((......))).....)))((((((((..((..(((((...))))))))))))))).)))))..)))))))))....))).................................((((.....)))).....)))).)))........))))....... ( -53.20)
>consensus
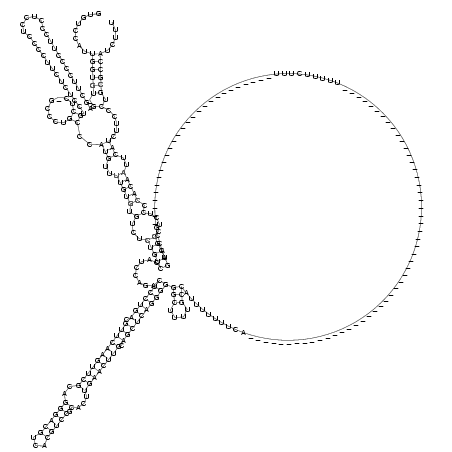
GUGUCCAUUGGUGU_GAGCUUCCCCUUCCCUCCUCCCCUUCUCUCCUGCUC_GCCCUGCCCAUGUUUUGUGUGUCUCUGUCCAUCCAGACUCCUGACGUUCAAGUUCGCAGGGACGUCACGUCCGCACUUGAACUUGCAGCUCAGGGGGGCUUUUGCCAUUUUUUUCA____________________________________________________UUUUUCUUU___________________________GCCAUGACGGCUCUCCCACAAUUCAUCUUCCCUGCGCCAUCUUU
........((((((.(...............................((........))..(((..(((((.(...(((((........(((((((.(((((((((((..((((((...))))).)...)))))))).))))))))))(((....))).......................................................................................................)))))....).)))))..))).....).))))))..... (-49.08 = -50.07 + 1.00)
481656_1.rnaz
RNAalifold consensus structure
Stefan Washietl
Last modified: Thu Aug 12 14:35:44 CEST 2004