Index >
Results for CNB 481656_0-rev
Input Alignment
CLUSTAL W(1.81) multiple sequence alignment
481656_ENSRNOG00000007049_RAT_9511_9952/1-454 ----NNNNNNNNNNNNNNAAGAAAAAAAAANNNNNNNNNNNNNNNNNNNNNNNNNNNNNN
481656_ENSG00000119866_HUMAN_9496_9947/1-454 TTAAGCNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN
481656_ENSMUSG00000000861_MOUSE_2861_3289/1-454 ---------NNNNNNNNNNNNAAGGAAAAA------AANNNNNNNNNNNNNNNNNNNNNN
481656_ENSDARG00000018362_ZEBRAFISH_9467_9847/1-454 ---------TTTTTTGAAAAGAGAGAGAGA------------------------------
481656_ENSRNOG00000007049_RAT_9511_9952/1-454 NNNNNNNNNNNNNNNNNNNNNNTGAAAAAAATGGCAAAAGCCCCCCTGAGCTGCAAGTTC
481656_ENSG00000119866_HUMAN_9496_9947/1-454 NNNNNNNNNNNNNNNNNNNNNNTGAAAAAAATGGCAAAAGCCCCCCTGAGCTGCAAGTTC
481656_ENSMUSG00000000861_MOUSE_2861_3289/1-454 NNNNNNNNNNNNNNNNNNNNNNTGAAAAAAATGGCAAAAGCCCCCCTGAGCTGCAAGTTC
481656_ENSDARG00000018362_ZEBRAFISH_9467_9847/1-454 ------------------GAGATG-AAAAAATGGCAAAAGCCCCCCTGAGCTGCAAGTTC
** ***********************************
481656_ENSRNOG00000007049_RAT_9511_9952/1-454 AAGTGCGGACGTGACGTCCCTGCGAACTTGAACGTCAGGAGTCTGGATGGACAGAGACAC
481656_ENSG00000119866_HUMAN_9496_9947/1-454 AAGTGCGGACGTGACGTCCCTGCGAACTTGAACGTCAGGAGTCTGGATGGACAGAGACAC
481656_ENSMUSG00000000861_MOUSE_2861_3289/1-454 AAGTGCGGACGTGACGTCCCTGCGAACTTGAACGTCAGGAGTCTGGATGGACAGAGACAC
481656_ENSDARG00000018362_ZEBRAFISH_9467_9847/1-454 AAGTGCGGACGTGACGTCCCTGCGAACTTGAACGTCAGGAG-ATGGATGGACACAGACAT
***************************************** ********** *****
481656_ENSRNOG00000007049_RAT_9511_9952/1-454 ACAAAACATGGGCAGGGC-GAGCAGGAGAGAAGGGGAGGAGGGAAGGGGAAGCTC-ACAC
481656_ENSG00000119866_HUMAN_9496_9947/1-454 ACAAAACATGGGCAGGGC-GAGCAGGAGAGAAGGGGAGGAGGGAAGGGGAAGCTC-ACAC
481656_ENSMUSG00000000861_MOUSE_2861_3289/1-454 ACAAAACATGGGCAGGGC-GAGCAGGAGAGAAGGGGAGGAGGGAAGGGGAAGCTC-ACAC
481656_ENSDARG00000018362_ZEBRAFISH_9467_9847/1-454 ACAGAACATGGGCAGGGCAAAGCCGTGGAGTA--GGAGGAGGGAGTGGAAATAACAAAAC
*** ************** *** * *** * ********** ** ** * * **
481656_ENSRNOG00000007049_RAT_9511_9952/1-454 CAATGGACACACATCAGGGGCTGGACATGAAAAAGAGACCAGGACAAGCCAATGGCCAGT
481656_ENSG00000119866_HUMAN_9496_9947/1-454 CAATGGACACACATCAGGGGCTGGACATGAAAAAGAGACCAGGACAAGCCAATGGCCAGT
481656_ENSMUSG00000000861_MOUSE_2861_3289/1-454 CAATGGACACACATCAGGGGCTGGACATGAAAAAGAGACCAGGACAAGCCAATGGCCAG-
481656_ENSDARG00000018362_ZEBRAFISH_9467_9847/1-454 CAATGGACACTCATTAGGGGCTGGACATGAAAAATAGACCCGAAGAAGCCAATGGACAG-
********** *** ******************* ***** * * ********** ***
481656_0-rev.aln
RNAz output
############################ RNAz 0.1 ##############################
Sequences: 4
Columns: 300
Mean pairwise identity: 79.27
Mean single sequence MFE: -62.25
Consensus MFE: -50.20
Energy contribution: -51.51
Covariance contribution: 1.31
Mean z-score: -2.14
Structure conservation index: 0.81
SVM decision value: 1.74
SVM RNA-class probability: 0.974945
Prediction: RNA
######################################################################
>481656_ENSRNOG00000007049_RAT_9511_9952/1-454
NNNNNNNNNNNNNNAAGAAAAAAAAANNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNUGAAAAAAAUGGCAAAAGCCCCCCUGAGCUGCAAGUUCAAGUGCGGACGUGACGUCCCUGCGAACUUGAACGUCAGGAGUCUGGAUGGACAGAGACACACAAAACAUGGGCAGGGCGAGCAGGAGAGAAGGGGAGGAGGGAAGGGGAAGCUCACACCAAUGGACACACAUCAGGGGCUGGACAUGAAAAAGAGACCAGGACAAGCCAAUGGCCAGU
.......................................................................................((((.....(((((((...((((..((((......))))....((.((((((...((((.....))))..((((......))))................)))))))).))))......))))).)).((...(((....)))...)).............((..(((((..(.((..........)))..).))))..)))))).. ( -62.50)
>481656_ENSG00000119866_HUMAN_9496_9947/1-454
UUAAGCNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNUGAAAAAAAUGGCAAAAGCCCCCCUGAGCUGCAAGUUCAAGUGCGGACGUGACGUCCCUGCGAACUUGAACGUCAGGAGUCUGGAUGGACAGAGACACACAAAACAUGGGCAGGGCGAGCAGGAGAGAAGGGGAGGAGGGAAGGGGAAGCUCACACCAAUGGACACACAUCAGGGGCUGGACAUGAAAAAGAGACCAGGACAAGCCAAUGGCCAGU
...........................................................................................((((.....(((((((...((((..((((......))))....((.((((((...((((.....))))..((((......))))................)))))))).))))......))))).)).((...(((....)))...)).............((..(((((..(.((..........)))..).))))..)))))).. ( -62.50)
>481656_ENSMUSG00000000861_MOUSE_2861_3289/1-454
NNNNNNNNNNNNAAGGAAAAAAANNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNUGAAAAAAAUGGCAAAAGCCCCCCUGAGCUGCAAGUUCAAGUGCGGACGUGACGUCCCUGCGAACUUGAACGUCAGGAGUCUGGAUGGACAGAGACACACAAAACAUGGGCAGGGCGAGCAGGAGAGAAGGGGAGGAGGGAAGGGGAAGCUCACACCAAUGGACACACAUCAGGGGCUGGACAUGAAAAAGAGACCAGGACAAGCCAAUGGCCAG
............................................................................((((.....(((((((...((((..((((......))))....((.((((((...((((.....))))..((((......))))................)))))))).))))......))))).)).((...(((....)))...)).............((..(((((..(.((..........)))..).))))..)))))). ( -62.50)
>481656_ENSDARG00000018362_ZEBRAFISH_9467_9847/1-454
UUUUUUGAAAAGAGAGAGAGAGAGAUGAAAAAAUGGCAAAAGCCCCCCUGAGCUGCAAGUUCAAGUGCGGACGUGACGUCCCUGCGAACUUGAACGUCAGGAGAUGGAUGGACACAGACAUACAGAACAUGGGCAGGGCAAAGCCGUGGAGUAGGAGGAGGGAGUGGAAAUAACAAAACCAAUGGACACUCAUUAGGGGCUGGACAUGAAAAAUAGACCCGAAGAAGCCAAUGGACAG
((((((....)))))).................((((...((((((((((..(..(..(((((((((((((((...)))))..))..))))))))(((.....(((..((....))..))).((.....)))))..(((...))))..)..))))......(((((....................)))))....))))))((...............))......))))........ ( -61.51)
>consensus
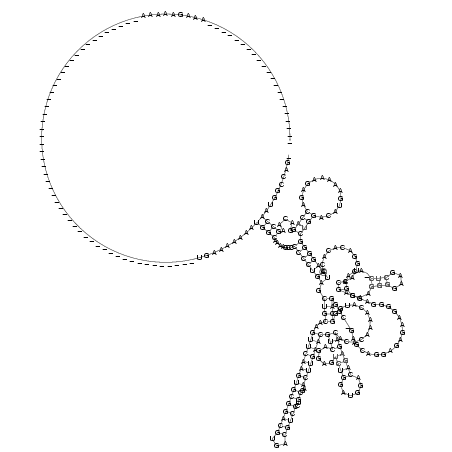
_____________________AAAGAAAAA____________________________________________________UGAAAAAAAUGGCAAAAGCCCCCCUGAGCUGCAAGUUCAAGUGCGGACGUGACGUCCCUGCGAACUUGAACGUCAGGAGUCUGGAUGGACAGAGACACACAAAACAUGGGCAGGGC_GAGCAGGAGAGAAGGGGAGGAGGGAAGGGGAAGCUC_ACACCAAUGGACACACAUCAGGGGCUGGACAUGAAAAAGAGACCAGGACAAGCCAAUGGCCAG_
...........................................................................................((((.......(((((((.((((..(((((((((((((((...)))))..))..))))))))(((.....((((......))))))).............)))).........................((...(((....)))....))............)))))))((((..............)))).....))))......... (-50.20 = -51.51 + 1.31)
481656_0-rev.rnaz
RNAalifold consensus structure
Stefan Washietl
Last modified: Thu Aug 12 14:35:44 CEST 2004