Index >
Results for CNB 461722-rev
Input Alignment
CLUSTAL W(1.81) multiple sequence alignment
461722_ENSMUSG00000034255_MOUSE_6286_6370/1-86 CACCTCTAACACATTCTTCTTGCTGGATTTGTCTTTGGGGGCCCAAGAGAGAGAGGCCCC
461722_ENSDARG00000006344_ZEBRAFISH_5690_5774/1-86 -ACCTCCAGAACGTTTTTCTTGCTTGATTTGTCCTTGGGCGCCCGGTTAATTGTGGCGCC
461722_SINFRUG00000131017_FUGU_9289_9374/1-86 AACCTCCAAAACATTCTTTTTGCTGGATTTGTCTTTTGTGGCCCAGCAAATAGTCGCTCC
461722_ENSRNOG00000028569_RAT_5899_5983/1-86 CACCTCTAGCACATTCTTCTTGCTGGATTTGTCTTTGGGGGCCCACGAGAGCGAGGCCCC
***** * ** ** ** ***** ******** ** * **** * * ** **
461722_ENSMUSG00000034255_MOUSE_6286_6370/1-86 CTTCAGTTCGACTGTGTATTCAGGG-
461722_ENSDARG00000006344_ZEBRAFISH_5690_5774/1-86 TTTCAGATCCACTGTGAATTCTGGGA
461722_SINFRUG00000131017_FUGU_9289_9374/1-86 TTTCAGGTCCACTGTGACCTCTGGGA
461722_ENSRNOG00000028569_RAT_5899_5983/1-86 CCTCAGTTCGACTGTATATTCAGGG-
**** ** ***** ** ***
461722-rev.aln
RNAz output
############################ RNAz 0.1 ##############################
Sequences: 4
Columns: 86
Mean pairwise identity: 76.89
Mean single sequence MFE: -26.90
Consensus MFE: -14.71
Energy contribution: -15.78
Covariance contribution: 1.06
Mean z-score: -2.03
Structure conservation index: 0.55
SVM decision value: 0.38
SVM RNA-class probability: 0.710848
Prediction: RNA
######################################################################
>461722_ENSMUSG00000034255_MOUSE_6286_6370/1-86
CACCUCUAACACAUUCUUCUUGCUGGAUUUGUCUUUGGGGGCCCAAGAGAGAGAGGCCCCCUUCAGUUCGACUGUGUAUUCAGGG
..(((...(((((.((.....((((((.........((((((((........).)))))))))))))..)).)))))....))). ( -28.20)
>461722_ENSDARG00000006344_ZEBRAFISH_5690_5774/1-86
ACCUCCAGAACGUUUUUCUUGCUUGAUUUGUCCUUGGGCGCCCGGUUAAUUGUGGCGCCUUUCAGAUCCACUGUGAAUUCUGGGA
...(((((((.....(((......((((((.....((((((((((....))).)))))))..))))))......)))))))))). ( -27.50)
>461722_SINFRUG00000131017_FUGU_9289_9374/1-86
AACCUCCAAAACAUUCUUUUUGCUGGAUUUGUCUUUUGUGGCCCAGCAAAUAGUCGCUCCUUUCAGGUCCACUGUGACCUCUGGGA
...............((.((((((((....(((......))))))))))).))....((((...(((((......)))))..)))) ( -22.90)
>461722_ENSRNOG00000028569_RAT_5899_5983/1-86
CACCUCUAGCACAUUCUUCUUGCUGGAUUUGUCUUUGGGGGCCCACGAGAGCGAGGCCCCCCUCAGUUCGACUGUAUAUUCAGGG
..(((((((((.........))))))).........((((((((.(....).).)))))))..(((.....)))........)). ( -29.00)
>consensus
CACCUCCAAAACAUUCUUCUUGCUGGAUUUGUCUUUGGGGGCCCAGGAAAGAGAGGCCCCCUUCAGUUCCACUGUGAAUUCAGGG_
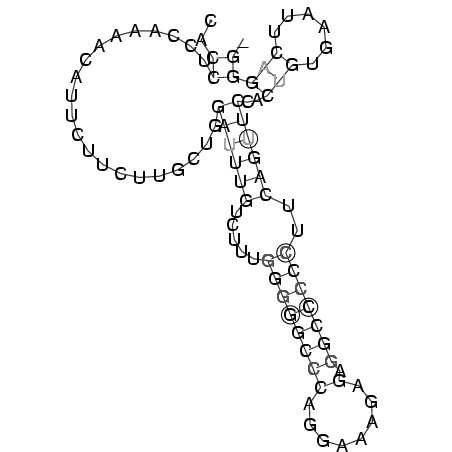
..((.....................((((((.....((((((((........).)))))))..))))))..(((......))))). (-14.71 = -15.78 + 1.06)
461722-rev.rnaz
RNAalifold consensus structure
Stefan Washietl
Last modified: Thu Aug 12 14:35:44 CEST 2004