Index >
Results for CNB 453983
Input Alignment
CLUSTAL W(1.81) multiple sequence alignment
453983_ENSMUSG00000007817_MOUSE_796_1001/1-207 -AGGTGTGTTCCTCTTTCCAGATGGGTCCCACCCAGGCGTATAACAGCCAATTCATGAAC
453983_ENSRNOG00000010488_RAT_796_1001/1-207 -AGGTGTGTTCCTCTTTCCAGATGGGTCCCACCCAGGCGTATAACAGCCAATTCATGAAC
453983_SINFRUG00000145892_FUGU_8334_8523/1-207 -AGATGAGCTCCTCTTTTCAGATGGCACCAAACCAGGCATACAACAGCCAGTTCATGAAC
453983_ENSDARG00000013677_ZEBRAFISH_9793_9998/1-207 CAGGTTTGCTCCTCATTCCAGATGGGGCCTACGCAAGGCTACAACAGCCAGTTTATGAAC
** * * ***** ** ******* ** * ** * ** ******** ** ******
453983_ENSMUSG00000007817_MOUSE_796_1001/1-207 CAACCCGGGCCTCGGGGGCCTGCCTCCATGGGGGGCAGCCTGAACCCTGCCGGCATGGCA
453983_ENSRNOG00000010488_RAT_796_1001/1-207 CAACCCGGGCCTCGGGGGCCTGCCTCCATGGGGGGCAGCATGAACCCTGCCAGCATGGCA
453983_SINFRUG00000145892_FUGU_8334_8523/1-207 CAGCCAGGGCCTCGCGGCCCCGCATC--------GCTCCCTGGA-------AACATGGGC
453983_ENSDARG00000013677_ZEBRAFISH_9793_9998/1-207 CAGCCAGGGCCTCGCGGACCCCCATCCATGGCAGGAGGCATGAACCCAGCTGCTATGGGT
** ** ******** ** ** * ** * * ** * ****
453983_ENSMUSG00000007817_MOUSE_796_1001/1-207 GCTGGCATGACACCCTCGGGCATGAGCGGCCCTCCCATGGGCATGAACCAGCCCCGACCA
453983_ENSRNOG00000010488_RAT_796_1001/1-207 GCTGGCATGACACCCTCGGGCATGAGCGGCCCTCCCATGGGCATGAACCAGCCCCGACCG
453983_SINFRUG00000145892_FUGU_8334_8523/1-207 GCAGGCATGAATGCGTCCAACATGAGTGGGCCTCCCATGGGAATGAACCAGCCCAGGGGC
453983_ENSDARG00000013677_ZEBRAFISH_9793_9998/1-207 GGTGGAATGAATAGCTCCAACATGAGCGGCCCACCGATGGGTATGAACCAGCCCAGAGGC
* ** **** ** ****** ** ** ** ***** ************ *
453983_ENSMUSG00000007817_MOUSE_796_1001/1-207 CCCGGCATCAGCCCCTTTGGCACACAC
453983_ENSRNOG00000010488_RAT_796_1001/1-207 CCTGGCATCAGCCCCTTTGGCACACAC
453983_SINFRUG00000145892_FUGU_8334_8523/1-207 CAAGGCATGGGGCCTTTTGGTGCCCA-
453983_ENSDARG00000013677_ZEBRAFISH_9793_9998/1-207 CCAGGCATGGGGCCCTTTGGTGGCCA-
* ***** * ** ***** **
//
453983.aln
RNAz output
############################ RNAz 0.1 ##############################
Sequences: 4
Columns: 207
Mean pairwise identity: 75.61
Mean single sequence MFE: -86.78
Consensus MFE: -44.54
Energy contribution: -51.73
Covariance contribution: 7.19
Mean z-score: -2.57
Structure conservation index: 0.51
SVM decision value: 1.61
SVM RNA-class probability: 0.967628
Prediction: RNA
######################################################################
>453983_ENSMUSG00000007817_MOUSE_796_1001/1-207
AGGUGUGUUCCUCUUUCCAGAUGGGUCCCACCCAGGCGUAUAACAGCCAAUUCAUGAACCAACCCGGGCCUCGGGGGCCUGCCUCCAUGGGGGGCAGCCUGAACCCUGCCGGCAUGGCAGCUGGCAUGACACCCUCGGGCAUGAGCGGCCCUCCCAUGGGCAUGAACCAGCCCCGACCACCCGGCAUCAGCCCCUUUGGCACACAC
..((((((........((...((((.....))))(((........))).................))(((..((((((.((((....((((((((.((((((..(.(((((((......))))))).)......)))))).......((((......))))........)))))..)))...))))...))))))..))))))))) ( -85.30)
>453983_ENSRNOG00000010488_RAT_796_1001/1-207
AGGUGUGUUCCUCUUUCCAGAUGGGUCCCACCCAGGCGUAUAACAGCCAAUUCAUGAACCAACCCGGGCCUCGGGGGCCUGCCUCCAUGGGGGGCAGCAUGAACCCUGCCAGCAUGGCAGCUGGCAUGACACCCUCGGGCAUGAGCGGCCCUCCCAUGGGCAUGAACCAGCCCCGACCGCCUGGCAUCAGCCCCUUUGGCACACAC
..(((((.........(((((.((((.....(((((((.......((...((((((........((((((.....))))))..((((((((((((..((((..((((((((((......))))))).((.....)))))))))....)))).))))))))))))))...))......))))))).....)))).)))))))))).. ( -90.22)
>453983_SINFRUG00000145892_FUGU_8334_8523/1-207
AGAUGAGCUCCUCUUUUCAGAUGGCACCAAACCAGGCAUACAACAGCCAGUUCAUGAACCAGCCAGGGCCUCGCGGCCCCGCAUCGCUCCCUGGAAACAUGGGCGCAGGCAUGAAUGCGUCCAACAUGAGUGGGCCUCCCAUGGGAAUGAACCAGCCCAGGGGCCAAGGCAUGGGGCCUUUUGGUGCCCA
.((.(((...)))...))....(((((((((..((((...((...(((.((((((...(((....(((((..(((....)))..(((((..((....))((((((((........))))))))....))))))))))....)))..))))))..(((....)))...))).))..))))))))))))).. ( -79.70)
>453983_ENSDARG00000013677_ZEBRAFISH_9793_9998/1-207
CAGGUUUGCUCCUCAUUCCAGAUGGGGCCUACGCAAGGCUACAACAGCCAGUUUAUGAACCAGCCAGGGCCUCGCGGACCCCCAUCCAUGGCAGGAGGCAUGAACCCAGCUGCUAUGGGUGGUGGAAUGAAUAGCUCCAACAUGAGCGGCCCACCGAUGGGUAUGAACCAGCCCAGAGGCCCAGGCAUGGGGCCCUUUGGUGGCCA
..((((...........((((..((((((.......((((.....))))..........((((((.(((((((.(((.((.(((((((((((((..((.......))..))))))))))))).))........((((......))))......))).(((((........)))))))))))).))).))))))))))))).)))). ( -91.90)
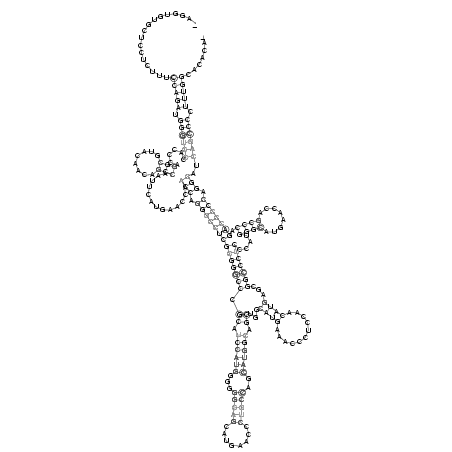
>consensus
_AGGUGUGCUCCUCUUUCCAGAUGGGUCCCACCCAGGCGUACAACAGCCAAUUCAUGAACCAACCAGGGCCUCGCGGGCCCGCAUCCAUGGGGGGCAGCAUGAACCCUGCCAGCAUGGCAGCUGGCAUGAAACCCUCCAACAUGAGCGGCCCUCCCAUGGGCAUGAACCAGCCCAGACCCCCAGGCAUCAGCCCCUUUGGCACACA_
.................(((((.((((((......(((........))).............(((.((((((((((((((.((.((((((...(((((........)))))..)))))).))...((((...........))))...)))))))....((((........)))).))))))).)))..)))))).)))))....... (-44.54 = -51.73 + 7.19)
453983.rnaz
RNAalifold consensus structure
Stefan Washietl
Last modified: Thu Aug 12 14:35:44 CEST 2004