Index >
Results for CNB 430453-rev
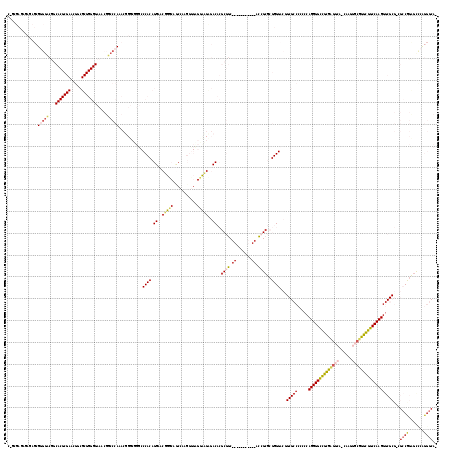
Input Alignment
CLUSTAL W(1.81) multiple sequence alignment
430453_ENSG00000114315_HUMAN_9189_9371/1-197 -CTCTATATATATCTGGGACTGCACGCGAACGGCTCGTGTGAAACTTCCCAAACTTTCTT
430453_SINFRUG00000139781_FUGU_9492_9685/1-197 T-TGTATTTATATCTCGCCCTGCACGCGAACGGCTCGTGTGAAACTTCCCAAACTTTCTT
430453_ENSMUSG00000022528_MOUSE_9551_9732/1-197 C-TCTATATATATCTGGGACTGCACGCGAACGGCTCGTGTGAAACTTCCCAAACTTTCTT
430453_ENSRNOG00000001720_RAT_9278_9459/1-197 C-TCTATATATATCTGGGACTGCACGCGAACGGCTCGTGTGAAACTTCCCAAACTTTCTT
430453_ENSDARG00000006514_ZEBRAFISH_9576_9769/1-197 T-CTTATTTATATCTCCGGCTGCACGCGAACGGCTCGTGTGAAACTTCCCAAACTTTCTT
*** ******* *****************************************
430453_ENSG00000114315_HUMAN_9189_9371/1-197 TCCCACAGTAACTTTCAGCCAATGGGAGGAGGAGACACGCGGC-----------ACCGCT
430453_SINFRUG00000139781_FUGU_9492_9685/1-197 TCCCACAGTAACTTTCCACCAATCAGGGAGAGGGACACGCGGCCCGA-GGGAGGACGGCT
430453_ENSMUSG00000022528_MOUSE_9551_9732/1-197 TCCCACAGTAACTTTCAGCCAATGGGAGGAAGAGACACGCGGC-----------ACCGCT
430453_ENSRNOG00000001720_RAT_9278_9459/1-197 TCCCACAGTAACTTTCAGCCAATGGGAGGAAGAGACACGCGGC-----------ACCGCT
430453_ENSDARG00000006514_ZEBRAFISH_9576_9769/1-197 TCCCACAGTAACTTTCTACCAATCAGCGCGCGGGACACGCGGCCCGAGGGGAGGATGGCT
**************** ***** * * * ********** * ***
430453_ENSG00000114315_HUMAN_9189_9371/1-197 CGTGGACCGCGCCCCCCCATTGGCCGCCAGGCA-CAAGGTCTGGCGGCCAATGGCGCG-C
430453_SINFRUG00000139781_FUGU_9492_9685/1-197 CGTGGTCGGCGCCCTCCCATTGGTTGTTTGACAGCTCGAGCTGGCGGCCAATGGCGCGAG
430453_ENSMUSG00000022528_MOUSE_9551_9732/1-197 CGTGGACTGCGCCCCCCCATTGGCCGCTAGGCA-CAAGGTCTGGCGGCCAATGGCGCG-C
430453_ENSRNOG00000001720_RAT_9278_9459/1-197 CGTGGACTGCGCCCCCCCATTGGCCGCCAGGCA-CAAGGTCTGGCGGCCAATGGCGCG-C
430453_ENSDARG00000006514_ZEBRAFISH_9576_9769/1-197 CGTGGTCGGCGCCCCTCCATTGGTTGTTTAGCG-TTTGAGCGGGTGGCCAATGGCGCG-C
***** * ****** ******* * * * * ** *************
430453_ENSG00000114315_HUMAN_9189_9371/1-197 GCCTGAGCCCAGGGCAC
430453_SINFRUG00000139781_FUGU_9492_9685/1-197 GCCCGGGTTCAGGGCA-
430453_ENSMUSG00000022528_MOUSE_9551_9732/1-197 GCCTGAGCCCGGGGCA-
430453_ENSRNOG00000001720_RAT_9278_9459/1-197 GCCTGAGCCCAGGGCA-
430453_ENSDARG00000006514_ZEBRAFISH_9576_9769/1-197 GGCCGGGTTTAGGGCAC
* * * * *****
430453-rev.aln
RNAz output
############################ RNAz 0.1 ##############################
Sequences: 5
Columns: 197
Mean pairwise identity: 82.38
Mean single sequence MFE: -79.64
Consensus MFE: -58.44
Energy contribution: -59.58
Covariance contribution: 1.14
Mean z-score: -2.41
Structure conservation index: 0.73
SVM decision value: 1.57
SVM RNA-class probability: 0.965022
Prediction: RNA
######################################################################
>430453_ENSG00000114315_HUMAN_9189_9371/1-197
CUCUAUAUAUAUCUGGGACUGCACGCGAACGGCUCGUGUGAAACUUCCCAAACUUUCUUUCCCACAGUAACUUUCAGCCAAUGGGAGGAGGAGACACGCGGCACCGCUCGUGGACCGCGCCCCCCCAUUGGCCGCCAGGCACAAGGUCUGGCGGCCAAUGGCGCGCGCCUGAGCCCAGGGCAC
...........((((((.((.(((((((.(((.(((((((...(((((...........(((((..((........))...))))))))))...)))))))..))).)))))...(((((....((((((((((((((((.....)))))))))))))))).)))))..)))))))))).... ( -83.00)
>430453_SINFRUG00000139781_FUGU_9492_9685/1-197
UUGUAUUUAUAUCUCGCCCUGCACGCGAACGGCUCGUGUGAAACUUCCCAAACUUUCUUUCCCACAGUAACUUUCCACCAAUCAGGGAGAGGGACACGCGGCCCGAGGGAGGACGGCUCGUGGUCGGCGCCCUCCCAUUGGUUGUUUGACAGCUCGAGCUGGCGGCCAAUGGCGCGAGGCCCGGGUUCAGGGCA
...............(((((((((((((.....)))))))......(((....(..(((((((.....................)))))))..)...(.((((((((((....((((.....))))...)))))(((((((((((....((((....)))))))))))))))...).))))))))..)))))). ( -79.50)
>430453_ENSMUSG00000022528_MOUSE_9551_9732/1-197
CUCUAUAUAUAUCUGGGACUGCACGCGAACGGCUCGUGUGAAACUUCCCAAACUUUCUUUCCCACAGUAACUUUCAGCCAAUGGGAGGAAGAGACACGCGGCACCGCUCGUGGACUGCGCCCCCCCAUUGGCCGCUAGGCACAAGGUCUGGCGGCCAAUGGCGCGCGCCUGAGCCCGGGGCA
...........((((((.((((.(((((.(((.(((((((...(((((...........(((((..((........))...))))))))))...)))))))..))).)))))....((((....((((((((((((((((.....)))))))))))))))).))))))...))))))))... ( -79.80)
>430453_ENSRNOG00000001720_RAT_9278_9459/1-197
CUCUAUAUAUAUCUGGGACUGCACGCGAACGGCUCGUGUGAAACUUCCCAAACUUUCUUUCCCACAGUAACUUUCAGCCAAUGGGAGGAAGAGACACGCGGCACCGCUCGUGGACUGCGCCCCCCCAUUGGCCGCCAGGCACAAGGUCUGGCGGCCAAUGGCGCGCGCCUGAGCCCAGGGCA
...........((((((.((((.(((((.(((.(((((((...(((((...........(((((..((........))...))))))))))...)))))))..))).)))))....((((....((((((((((((((((.....)))))))))))))))).))))))...))))))))... ( -82.80)
>430453_ENSDARG00000006514_ZEBRAFISH_9576_9769/1-197
UCUUAUUUAUAUCUCCGGCUGCACGCGAACGGCUCGUGUGAAACUUCCCAAACUUUCUUUCCCACAGUAACUUUCUACCAAUCAGCGCGCGGGACACGCGGCCCGAGGGGAGGAUGGCUCGUGGUCGGCGCCCCUCCAUUGGUUGUUUAGCGUUUGAGCGGGUGGCCAAUGGCGCGCGGCCGGGUUUAGGGCAC
.........((..(((((((((.(((....(((.((((((.....((((.................(((......)))......(....)))))))))))))).((((((....((((.....))))...))))))(((((((..(((.((......)))))..)))))))))).)))))))))..))...... ( -73.10)
>consensus
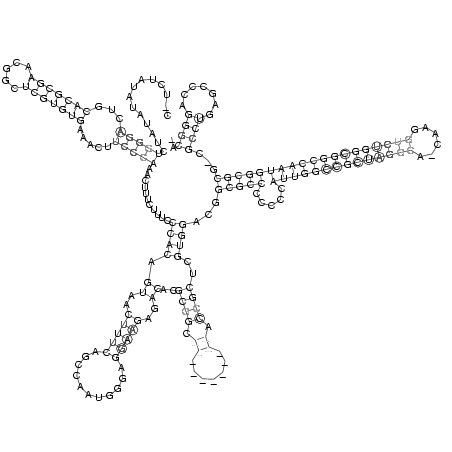
C_UCUAUAUAUAUCUGGGACUGCACGCGAACGGCUCGUGUGAAACUUCCCAAACUUUCUUUCCCACAGUAACUUUCAGCCAAUGGGAGGAAGAGACACGCGGC___________ACCGCUCGUGGACGGCGCCCCCCCAUUGGCCGCUAGGCA_CAAGGUCUGGCGGCCAAUGGCGCG_CGCCUGAGCCCAGGGCA_
..............(((((...(((((((.....))))))).....)))))...........((((.((..(((((...........)))))..))..((((.((.......)).))))..))))...(((((.....((((((((((((((......)))))))))))))))))))...((((.......)))).. (-58.44 = -59.58 + 1.14)
430453-rev.rnaz
RNAalifold consensus structure
Stefan Washietl
Last modified: Thu Aug 12 14:35:44 CEST 2004