Index >
Results for CNB 430423-rev
Input Alignment
CLUSTAL W(1.81) multiple sequence alignment
430423_ENSG00000114315_HUMAN_9189_9369/1-194 CTATATATATCTGGGACTGCACGCGAACGGCTCGTGTGAAACTTCCCAAACTTTCTTTCC
430423_ENSMUSG00000022528_MOUSE_9550_9730/1-194 CTATATATATCTGGGACTGCACGCGAACGGCTCGTGTGAAACTTCCCAAACTTTCTTTCC
430423_ENSDARG00000006514_ZEBRAFISH_9576_9766/1-194 -TATTTATATCTCCGGCTGCACGCGAACGGCTCGTGTGAAACTTCCCAAACTTTCTTTCC
430423_SINFRUG00000139781_FUGU_9492_9683/1-194 GTATTTATATCTCGCCCTGCACGCGAACGGCTCGTGTGAAACTTCCCAAACTTTCTTTCC
*** ******* ********************************************
430423_ENSG00000114315_HUMAN_9189_9369/1-194 CACAGTAACTTTCAGCCAATGGGAGGAGGAGACACGCGGC-----------ACCGCTCGT
430423_ENSMUSG00000022528_MOUSE_9550_9730/1-194 CACAGTAACTTTCAGCCAATGGGAGGAAGAGACACGCGGC-----------ACCGCTCGT
430423_ENSDARG00000006514_ZEBRAFISH_9576_9766/1-194 CACAGTAACTTTCTACCAATCAGCGCGCGGGACACGCGGCCCGAGGGGAGGATGGCTCGT
430423_SINFRUG00000139781_FUGU_9492_9683/1-194 CACAGTAACTTTCCACCAATCAGGGAGAGGGACACGCGGCCCGA-GGGAGGACGGCTCGT
************* ***** * * * ********** * ******
430423_ENSG00000114315_HUMAN_9189_9369/1-194 GGACCGCGCCCCCCCATTGGCCGCCAGGCA-CAAGGTCTGGCGGCCAATGGCGCG-CGCC
430423_ENSMUSG00000022528_MOUSE_9550_9730/1-194 GGACTGCGCCCCCCCATTGGCCGCTAGGCA-CAAGGTCTGGCGGCCAATGGCGCG-CGCC
430423_ENSDARG00000006514_ZEBRAFISH_9576_9766/1-194 GGTCGGCGCCCCTCCATTGGTTGTTTAGCG-TTTGAGCGGGTGGCCAATGGCGCG-CGGC
430423_SINFRUG00000139781_FUGU_9492_9683/1-194 GGTCGGCGCCCTCCCATTGGTTGTTTGACAGCTCGAGCTGGCGGCCAATGGCGCGAGGCC
** * ****** ******* * * * * ** ************* * *
430423_ENSG00000114315_HUMAN_9189_9369/1-194 TGAGCCCAGGGCAC
430423_ENSMUSG00000022528_MOUSE_9550_9730/1-194 TGAGCCCGGGGCAC
430423_ENSDARG00000006514_ZEBRAFISH_9576_9766/1-194 CGGGTTTAGGGCAC
430423_SINFRUG00000139781_FUGU_9492_9683/1-194 CGGGTTCAGGGCA-
* * *****
430423-rev.aln
RNAz output
############################ RNAz 0.1 ##############################
Sequences: 4
Columns: 194
Mean pairwise identity: 80.79
Mean single sequence MFE: -78.85
Consensus MFE: -54.41
Energy contribution: -56.22
Covariance contribution: 1.81
Mean z-score: -2.01
Structure conservation index: 0.69
SVM decision value: 0.92
SVM RNA-class probability: 0.880959
Prediction: RNA
######################################################################
>430423_ENSG00000114315_HUMAN_9189_9369/1-194
CUAUAUAUAUCUGGGACUGCACGCGAACGGCUCGUGUGAAACUUCCCAAACUUUCUUUCCCACAGUAACUUUCAGCCAAUGGGAGGAGGAGACACGCGGCACCGCUCGUGGACCGCGCCCCCCCAUUGGCCGCCAGGCACAAGGUCUGGCGGCCAAUGGCGCGCGCCUGAGCCCAGGGCAC
.........((((((.((.(((((((.(((.(((((((...(((((...........(((((..((........))...))))))))))...)))))))..))).)))))...(((((....((((((((((((((((.....)))))))))))))))).)))))..)))))))))).... ( -83.00)
>430423_ENSMUSG00000022528_MOUSE_9550_9730/1-194
CUAUAUAUAUCUGGGACUGCACGCGAACGGCUCGUGUGAAACUUCCCAAACUUUCUUUCCCACAGUAACUUUCAGCCAAUGGGAGGAAGAGACACGCGGCACCGCUCGUGGACUGCGCCCCCCCAUUGGCCGCUAGGCACAAGGUCUGGCGGCCAAUGGCGCGCGCCUGAGCCCGGGGCAC
.........((((((.((((.(((((.(((.(((((((...(((((...........(((((..((........))...))))))))))...)))))))..))).)))))....((((....((((((((((((((((.....)))))))))))))))).))))))...)))))))).... ( -79.80)
>430423_ENSDARG00000006514_ZEBRAFISH_9576_9766/1-194
UAUUUAUAUCUCCGGCUGCACGCGAACGGCUCGUGUGAAACUUCCCAAACUUUCUUUCCCACAGUAACUUUCUACCAAUCAGCGCGCGGGACACGCGGCCCGAGGGGAGGAUGGCUCGUGGUCGGCGCCCCUCCAUUGGUUGUUUAGCGUUUGAGCGGGUGGCCAAUGGCGCGCGGCCGGGUUUAGGGCAC
......((..(((((((((.(((....(((.((((((.....((((.................(((......)))......(....)))))))))))))).((((((....((((.....))))...))))))(((((((..(((.((......)))))..)))))))))).)))))))))..))...... ( -73.10)
>430423_SINFRUG00000139781_FUGU_9492_9683/1-194
GUAUUUAUAUCUCGCCCUGCACGCGAACGGCUCGUGUGAAACUUCCCAAACUUUCUUUCCCACAGUAACUUUCCACCAAUCAGGGAGAGGGACACGCGGCCCGAGGGAGGACGGCUCGUGGUCGGCGCCCUCCCAUUGGUUGUUUGACAGCUCGAGCUGGCGGCCAAUGGCGCGAGGCCCGGGUUCAGGGCA
.............(((((((((((((.....)))))))......(((....(..(((((((.....................)))))))..)...(.((((((((((....((((.....))))...)))))(((((((((((....((((....)))))))))))))))...).))))))))..)))))). ( -79.50)
>consensus
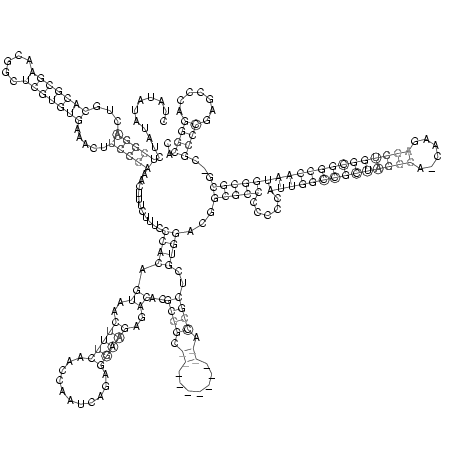
CUAUAUAUAUCUCGGACUGCACGCGAACGGCUCGUGUGAAACUUCCCAAACUUUCUUUCCCACAGUAACUUUCAACCAAUCAGAGGAAGAGACACGCGGC___________ACCGCUCGUGGACGGCGCCCCCCCAUUGGCCGCUAGGCA_CAAGAGCUGGCGGCCAAUGGCGCG_CGCCCGAGCCCAGGGCAC
...........(((((...(((((((.....))))))).....)))))...........((((.((..(((((...........)))))..))..((((.((.......)).))))..))))...(((((.....((((((((((((((......)))))))))))))))))))...((((.......)))).. (-54.41 = -56.22 + 1.81)
430423-rev.rnaz
RNAalifold consensus structure
Stefan Washietl
Last modified: Thu Aug 12 14:35:44 CEST 2004