Index >
Results for CNB 414286-rev
Input Alignment
CLUSTAL W(1.81) multiple sequence alignment
414286_ENSG00000140416_HUMAN_5571_5706/1-137 TTACATCGGTGGCCTTTTTCTCTGCCAGCTCCAGCTTCTCCTGGGCATCTTTGAGAGCCT
414286_SINFRUG00000130484_FUGU_7336_7470/1-137 -TACGTCTGTGGCTTTCTTTTCTGCCAGTTCAAGCTTCTCCTGAGCATCTTTAAGAGCCT
414286_ENSRNOG00000018184_RAT_5838_5973/1-137 TTACATCTGTGGCCTTTTTCTCCGCCAGCTCCAGTTTCTCCTGGGCATCTTTGAGAGCCT
414286_ENSMUSG00000032366_MOUSE_6042_6176/1-137 -TACATCTGTGGCCTTTTTCTCCGCCAGCTCCAGTTTCTCCTGGGCATCTTTGAGAGCCT
*** ** ***** ** ** ** ***** ** ** ******** ******** *******
414286_ENSG00000140416_HUMAN_5571_5706/1-137 CAGAGTATTTGTCCAGTTCATCTTCGGTGCCCTTGAGTTTCTTTTGCAGTGACACCAGCT
414286_SINFRUG00000130484_FUGU_7336_7470/1-137 CGGAGAACTTGTCCAACTCATCTTCTGTCCCCTTTAATTTCTTCTGCAGGGCTACCAAAT
414286_ENSRNOG00000018184_RAT_5838_5973/1-137 CGGAGTATTTGTCCAGTTCATCTTCAGTGCCCTTGAGTTTCTTTTGCAGTGACACCAGCT
414286_ENSMUSG00000032366_MOUSE_6042_6176/1-137 CGGAGTATTTGTCCAGTTCATCTTCAGTGCCCTTGAGTTTCTTTTGCAGTGACACCAGCT
* *** * ******* ******** ** ***** * ****** ***** * **** *
414286_ENSG00000140416_HUMAN_5571_5706/1-137 CATCTTCCAGCTGGG-A
414286_SINFRUG00000130484_FUGU_7336_7470/1-137 CGTCCTCAAGCTGAGG-
414286_ENSRNOG00000018184_RAT_5838_5973/1-137 CATCTTCCAGCTGGG-A
414286_ENSMUSG00000032366_MOUSE_6042_6176/1-137 CATCTTCCAGCTGGG-A
* ** ** ***** *
414286-rev.aln
RNAz output
############################ RNAz 0.1 ##############################
Sequences: 4
Columns: 137
Mean pairwise identity: 87.29
Mean single sequence MFE: -36.22
Consensus MFE: -25.01
Energy contribution: -26.08
Covariance contribution: 1.06
Mean z-score: -1.46
Structure conservation index: 0.69
SVM decision value: -0.05
SVM RNA-class probability: 0.509414
Prediction: RNA
######################################################################
>414286_ENSG00000140416_HUMAN_5571_5706/1-137
UUACAUCGGUGGCCUUUUUCUCUGCCAGCUCCAGCUUCUCCUGGGCAUCUUUGAGAGCCUCAGAGUAUUUGUCCAGUUCAUCUUCGGUGCCCUUGAGUUUCUUUUGCAGUGACACCAGCUCAUCUUCCAGCUGGGA
((((...((..(.........)..)).((...((...(((..(((((((...((((((..((((...))))....)))).))...)))))))..)))...))...)).))))..((((((........)))))).. ( -38.40)
>414286_SINFRUG00000130484_FUGU_7336_7470/1-137
UACGUCUGUGGCUUUCUUUUCUGCCAGUUCAAGCUUCUCCUGAGCAUCUUUAAGAGCCUCGGAGAACUUGUCCAACUCAUCUUCUGUCCCCUUUAAUUUCUUCUGCAGGGCUACCAAAUCGUCCUCAAGCUGAGG
........((((..........))))(..((((.((((((.(((..((.....))..)))))))))))))..)..........................((((.(((((((.........)))))...)).)))) ( -30.90)
>414286_ENSRNOG00000018184_RAT_5838_5973/1-137
UUACAUCUGUGGCCUUUUUCUCCGCCAGCUCCAGUUUCUCCUGGGCAUCUUUGAGAGCCUCGGAGUAUUUGUCCAGUUCAUCUUCAGUGCCCUUGAGUUUCUUUUGCAGUGACACCAGCUCAUCUUCCAGCUGGGA
((((.....((((..........))))((...((...(((..((((((....((((((...(((.......))).)))).))....))))))..)))...))...)).))))..((((((........)))))).. ( -38.30)
>414286_ENSMUSG00000032366_MOUSE_6042_6176/1-137
UACAUCUGUGGCCUUUUUCUCCGCCAGCUCCAGUUUCUCCUGGGCAUCUUUGAGAGCCUCGGAGUAUUUGUCCAGUUCAUCUUCAGUGCCCUUGAGUUUCUUUUGCAGUGACACCAGCUCAUCUUCCAGCUGGGA
(((.....((((..........))))((...((...(((..((((((....((((((...(((.......))).)))).))....))))))..)))...))...)).)))...((((((........)))))).. ( -37.30)
>consensus
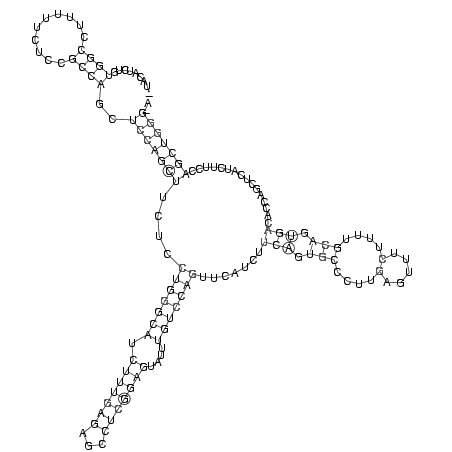
_UACAUCUGUGGCCUUUUUCUCCGCCAGCUCCAGCUUCUCCUGGGCAUCUUUGAGAGCCUCGGAGUAUUUGUCCAGUUCAUCUUCAGUGCCCUUGAGUUUCUUUUGCAGUGACACCAGCUCAUCUUCCAGCUGGG_A
.........((((..........))))..(((((((....(((((((.(((((((...)))))))....))))))).......(((.(((....(.....)....))).)))................))))))).. (-25.01 = -26.08 + 1.06)
414286-rev.rnaz
RNAalifold consensus structure
Stefan Washietl
Last modified: Thu Aug 12 14:35:44 CEST 2004