Index >
Results for CNB 393786
Input Alignment
CLUSTAL W(1.81) multiple sequence alignment
393786_ENSG00000138449_HUMAN_8851_8915/1-67 TAAGGCTT-TGCCTTTCCAACTTCAGCTACAGTGTTAGCTAAGTTTGGAAAGAAG-GAAA
393786_ENSDARG00000000241_ZEBRAFISH_9810_9876/1-67 GAAAGGTTATTTCTCTCCGACTTCAGCTACAGTGATAGCTAAGTTTGGAGAGGAGAGAAG
393786_ENSMUSG00000025993_MOUSE_9754_9818/1-67 TAAGGCTT-TGGCTTTCCAACTTCAGCTACAGTGTTAGCTAAGTTTGGAAAGAAG-ACAA
393786_SINFRUG00000122977_FUGU_3359_3421/1-67 -AAAAGCA-TTTCTTTCCAACTTCAGCTACAGTGTTAGCTAAGTTTGGAGGGGAG-GAAA
393786_ENSRNOG00000003872_RAT_9703_9767/1-67 TAAGGCTT-TAGCTTTCCAACTTCAGCTACAGTGTTAGCTAAGTTTGGAAAGAAG-ACAT
** * ** *** *************** ************** * ** *
393786_ENSG00000138449_HUMAN_8851_8915/1-67 AAAGAAA
393786_ENSDARG00000000241_ZEBRAFISH_9810_9876/1-67 GGAGATA
393786_ENSMUSG00000025993_MOUSE_9754_9818/1-67 AAAGAAG
393786_SINFRUG00000122977_FUGU_3359_3421/1-67 ACGGGA-
393786_ENSRNOG00000003872_RAT_9703_9767/1-67 AAAGAAG
*
//
393786.aln
RNAz output
############################ RNAz 0.1 ##############################
Sequences: 5
Columns: 67
Mean pairwise identity: 79.79
Mean single sequence MFE: -18.86
Consensus MFE: -17.60
Energy contribution: -16.88
Covariance contribution: -0.72
Mean z-score: -3.27
Structure conservation index: 0.93
SVM decision value: 3.15
SVM RNA-class probability: 0.998586
Prediction: RNA
######################################################################
>393786_ENSG00000138449_HUMAN_8851_8915/1-67
UAAGGCUUUGCCUUUCCAACUUCAGCUACAGUGUUAGCUAAGUUUGGAAAGAAGGAAAAAAGAAA
.....((((.((((((((((((.(((((......)))))))).)))))...))))...))))... ( -17.40)
>393786_ENSDARG00000000241_ZEBRAFISH_9810_9876/1-67
GAAAGGUUAUUUCUCUCCGACUUCAGCUACAGUGAUAGCUAAGUUUGGAGAGGAGAGAAGGGAGAUA
.........((((((((((((((.(((((......)))))))).)))))))))))............ ( -21.50)
>393786_ENSMUSG00000025993_MOUSE_9754_9818/1-67
UAAGGCUUUGGCUUUCCAACUUCAGCUACAGUGUUAGCUAAGUUUGGAAAGAAGACAAAAAGAAG
......((((.(((((((((((.(((((......)))))))).))))))))....))))...... ( -18.40)
>393786_SINFRUG00000122977_FUGU_3359_3421/1-67
AAAAGCAUUUCUUUCCAACUUCAGCUACAGUGUUAGCUAAGUUUGGAGGGGAGGAAAACGGGA
.......((((((((((((((.(((((......)))))))).))))))))))).......... ( -18.50)
>393786_ENSRNOG00000003872_RAT_9703_9767/1-67
UAAGGCUUUAGCUUUCCAACUUCAGCUACAGUGUUAGCUAAGUUUGGAAAGAAGACAUAAAGAAG
.....(((((.(((((((((((.(((((......)))))))).))))))))......)))))... ( -18.50)
>consensus
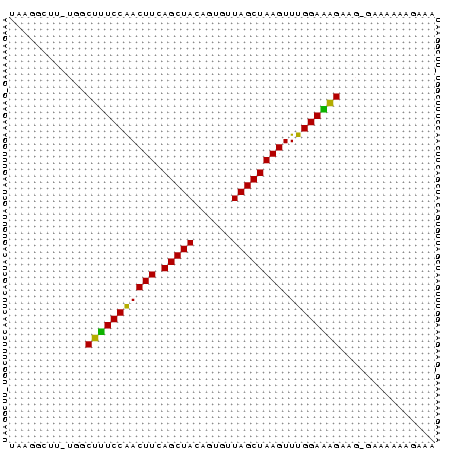
UAAGGCUU_UGGCUUUCCAACUUCAGCUACAGUGUUAGCUAAGUUUGGAAAGAAG_GAAAAAAGAAA
............(((((((((((.(((((......)))))))).))))))))............... (-17.60 = -16.88 + -0.72)
393786.rnaz
RNAalifold consensus structure
Stefan Washietl
Last modified: Thu Aug 12 14:35:44 CEST 2004