Index >
Results for CNB 386626
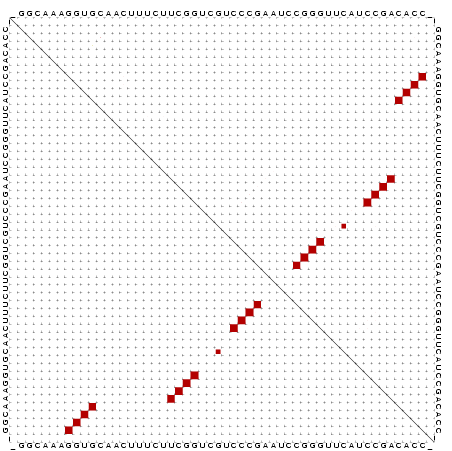
Input Alignment
CLUSTAL W(1.81) multiple sequence alignment
386626_ENSG00000158066_HUMAN_936_989/1-54 AGGCCAAGGTGCAACTTCCTTCGGTCGTCCCGAATCCGGGTTCATCCGACACCA
386626_ENSMUSG00000038900_MOUSE_936_989/1-54 AGGCAACGGTGCAACTTTCTTCGGTCGTCCCGAATCCGGGTTCATCCGACACCA
386626_ENSDARG00000006691_ZEBRAFISH_905_956/1-54 -GGTGAAGGTGCAACAGTTTTCGGTCGTCCCGAATCCGGGTTCATCCGACACC-
386626_SINFRUG00000134705_FUGU_6362_6413/1-54 -GGTGAAGGTGCAACAGTAATCGGTCGTCCCGACTGCGGGTTCATCCGACACC-
386626_ENSRNOG00000016220_RAT_9891_9942/1-54 -GGCAACGGTGCAACTTTCTTCGGTCGTCCCGAATCCGGGTTCATCCGACACC-
** * ******** ************* * *****************
//
386626.aln
RNAz output
############################ RNAz 0.1 ##############################
Sequences: 5
Columns: 54
Mean pairwise identity: 87.45
Mean single sequence MFE: -16.88
Consensus MFE: -14.98
Energy contribution: -14.98
Covariance contribution: 0.00
Mean z-score: -1.33
Structure conservation index: 0.89
SVM decision value: -0.02
SVM RNA-class probability: 0.523598
Prediction: RNA
######################################################################
>386626_ENSG00000158066_HUMAN_936_989/1-54
AGGCCAAGGUGCAACUUCCUUCGGUCGUCCCGAAUCCGGGUUCAUCCGACACCA
.......((((.........((((..(.((((....))))..)..)))))))). ( -16.00)
>386626_ENSMUSG00000038900_MOUSE_936_989/1-54
AGGCAACGGUGCAACUUUCUUCGGUCGUCCCGAAUCCGGGUUCAUCCGACACCA
.(....)((((.........((((..(.((((....))))..)..)))))))). ( -17.30)
>386626_ENSDARG00000006691_ZEBRAFISH_905_956/1-54
GGUGAAGGUGCAACAGUUUUCGGUCGUCCCGAAUCCGGGUUCAUCCGACACC
((((..((((.(((.(..(((((.....)))))..)..))))).))..)))) ( -17.50)
>386626_SINFRUG00000134705_FUGU_6362_6413/1-54
GGUGAAGGUGCAACAGUAAUCGGUCGUCCCGACUGCGGGUUCAUCCGACACC
((((..((((.(((.(((.((((.....)))).)))..))))).))..)))) ( -18.20)
>386626_ENSRNOG00000016220_RAT_9891_9942/1-54
GGCAACGGUGCAACUUUCUUCGGUCGUCCCGAAUCCGGGUUCAUCCGACACC
(....)((((.........((((..(.((((....))))..)..)))))))) ( -15.40)
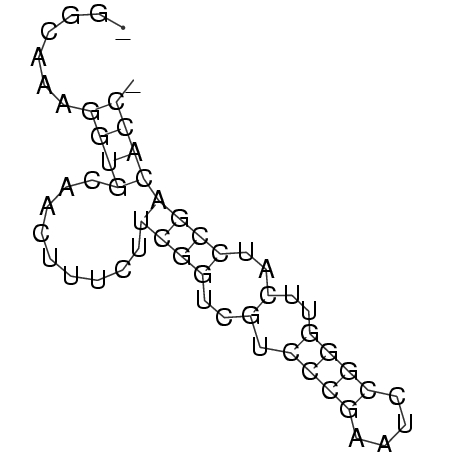
>consensus
_GGCAAAGGUGCAACUUUCUUCGGUCGUCCCGAAUCCGGGUUCAUCCGACACC_
.......((((.........((((..(.((((....))))..)..)))))))). (-14.98 = -14.98 + 0.00)
386626.rnaz
RNAalifold consensus structure
Stefan Washietl
Last modified: Thu Aug 12 14:35:44 CEST 2004