Index >
Results for CNB 363129
Input Alignment
CLUSTAL W(1.81) multiple sequence alignment
363129_ENSG00000104313_HUMAN_8175_8288/1-119 C-CCCCCTGACAAGGGGAGG--AGGAGGATAGACCTGCA-GGAGGAAACAACAGTTGAGA
363129_ENSDARG00000014259_ZEBRAFISH_7973_8070/1-119 ---------------GGAGG--AGG--GA-AGACCTGGAAGGAGGAAACAACGGTGGAGG
363129_ENSMUSG00000025932_MOUSE_8198_8313/1-119 C-CCTCCTGACAAGGGGAGGGGAGGAGGATAGACCTGCA-GGAAGAAACAACAGTTGAGC
363129_ENSRNOG00000007590_RAT_8238_8352/1-119 -TCCTCTTGACAAGGGGAGGGGAGGAGGAGAGACCTGCA-GGAAGAAACAGCAGTTGAGC
***** *** ** ******* * *** ****** * ** ***
363129_ENSG00000104313_HUMAN_8175_8288/1-119 CCTC-AACTTAAAGACAATATGCCTTTCACTGCTGCAGTATCCTCCTTTTTCTCTTTTG
363129_ENSDARG00000014259_ZEBRAFISH_7973_8070/1-119 GGTTGAACCTACAGACAATGTGCCTCTCTGTTCCACCGACTCCTC-TCTTCCCCTTCTG
363129_ENSMUSG00000025932_MOUSE_8198_8313/1-119 CCTC-CACTTAAAGACACTATGCCTTTCCTGGCTGCAGTATCCTCCCTTTTCTCGTTTG
363129_ENSRNOG00000007590_RAT_8238_8352/1-119 CCTC-CACTTAAAGATACTATGCCTTTCCTGGCCGCAGTATCCTCCCTTTTCTCTTTT-
* ** ** *** * * ***** ** * * * ***** ** * * * *
//
363129.aln
RNAz output
############################ RNAz 0.1 ##############################
Sequences: 4
Columns: 119
Mean pairwise identity: 73.50
Mean single sequence MFE: -40.10
Consensus MFE: -16.38
Energy contribution: -17.88
Covariance contribution: 1.50
Mean z-score: -2.58
Structure conservation index: 0.41
SVM decision value: 0.56
SVM RNA-class probability: 0.783159
Prediction: RNA
######################################################################
>363129_ENSG00000104313_HUMAN_8175_8288/1-119
CCCCCCUGACAAGGGGAGGAGGAGGAUAGACCUGCAGGAGGAAACAACAGUUGAGACCUCAACUUAAAGACAAUAUGCCUUUCACUGCUGCAGUAUCCUCCUUUUUCUCUUUUG
(((((((....))))).))(((((((((...((((((.((........((((((....)))))).((((.(.....).))))..)).)))))))))))))))............ ( -42.80)
>363129_ENSDARG00000014259_ZEBRAFISH_7973_8070/1-119
GGAGGAGGGAAGACCUGGAAGGAGGAAACAACGGUGGAGGGGUUGAACCUACAGACAAUGUGCCUCUCUGUUCCACCGACUCCUCUCUUCCCCUUCUG
((((((((.....)))((((((((((.....((((((((.((..((...((((.....))))..)).)).))))))))..))))).)))))))))).. ( -40.90)
>363129_ENSMUSG00000025932_MOUSE_8198_8313/1-119
CCCUCCUGACAAGGGGAGGGGAGGAGGAUAGACCUGCAGGAAGAAACAACAGUUGAGCCCUCCACUUAAAGACACUAUGCCUUUCCUGGCUGCAGUAUCCUCCCUUUUCUCGUUUG
(((((((......)))))))..((((((((...((((((((.(.(((....)))...)..)))...............(((......))))))))))))))))............. ( -38.80)
>363129_ENSRNOG00000007590_RAT_8238_8352/1-119
UCCUCUUGACAAGGGGAGGGGAGGAGGAGAGACCUGCAGGAAGAAACAGCAGUUGAGCCCUCCACUUAAAGAUACUAUGCCUUUCCUGGCCGCAGUAUCCUCCCUUUUCUCUUUU
((((((.....))))))((((((((((.(((..((((.(.......).))))(((((.......))))).((((((..(((......)))...)))))))))))))))))))... ( -37.90)
>consensus
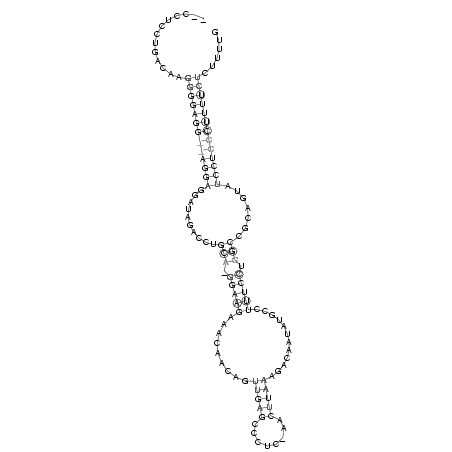
__CCUCCUGACAAGGGGAGG__AGGAGGAUAGACCUGCA_GGAAGAAACAACAGUUGAGCCCUC_AACUUAAAGACAAUAUGCCUUUCCUGGCCGCAGUAUCCUCCCUUUUCUCUUUUG
.............(((((((((((((..........(((.(((((.........(((((........)))))............))))).))).......)))))))))))))...... (-16.38 = -17.88 + 1.50)
363129.rnaz
RNAalifold consensus structure
Stefan Washietl
Last modified: Thu Aug 12 14:35:44 CEST 2004