Index >
Results for CNB 363120
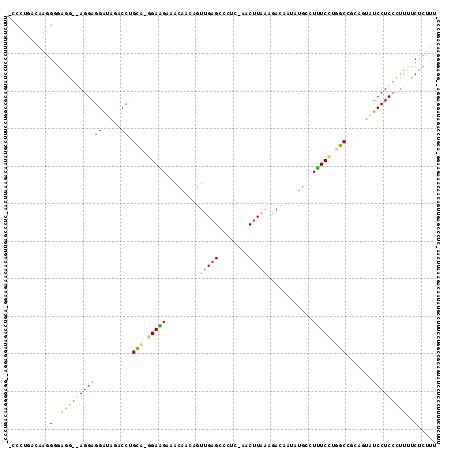
Input Alignment
CLUSTAL W(1.81) multiple sequence alignment
363120_ENSG00000104313_HUMAN_8178_8286/1-114 -CCCTGACAAGGGGAGG--AGGAGGATAGACCTGCA-GGAGGAAACAACAGTTGAGACCT
363120_ENSDARG00000014259_ZEBRAFISH_7958_8067/1-114 TCCGTCATTGGGCCGGG--AGGAGGGAAGACCTGGAAGGAGGAAACAACGGTGGAGGGGT
363120_ENSMUSG00000025932_MOUSE_8201_8311/1-114 -TCCTGACAAGGGGAGGGGAGGAGGATAGACCTGCA-GGAAGAAACAACAGTTGAGCCCT
363120_ENSRNOG00000007590_RAT_8241_8351/1-114 -TCTTGACAAGGGGAGGGGAGGAGGAGAGACCTGCA-GGAAGAAACAGCAGTTGAGCCCT
* * * ** ** ****** ******* * *** ****** * ** *** *
363120_ENSG00000104313_HUMAN_8178_8286/1-114 C-AACTTAAAGACAATATGCCTTTCACTGCTGCAGTATCCTCCTTTTTCTCTTT
363120_ENSDARG00000014259_ZEBRAFISH_7958_8067/1-114 TGAACCTACAGACAATGTGCCTCTCTGTTCCACCGACTCCTC-TCTTCCCCTT-
363120_ENSMUSG00000025932_MOUSE_8201_8311/1-114 C-CACTTAAAGACACTATGCCTTTCCTGGCTGCAGTATCCTCCCTTTTCTCGTT
363120_ENSRNOG00000007590_RAT_8241_8351/1-114 C-CACTTAAAGATACTATGCCTTTCCTGGCCGCAGTATCCTCCCTTTTCTCTTT
** ** *** * * ***** ** * * * ***** ** * * *
//
363120.aln
RNAz output
############################ RNAz 0.1 ##############################
Sequences: 4
Columns: 114
Mean pairwise identity: 75.78
Mean single sequence MFE: -39.90
Consensus MFE: -19.66
Energy contribution: -20.23
Covariance contribution: 0.56
Mean z-score: -2.48
Structure conservation index: 0.49
SVM decision value: 1.08
SVM RNA-class probability: 0.912551
Prediction: RNA
######################################################################
>363120_ENSG00000104313_HUMAN_8178_8286/1-114
CCCUGACAAGGGGAGGAGGAGGAUAGACCUGCAGGAGGAAACAACAGUUGAGACCUCAACUUAAAGACAAUAUGCCUUUCACUGCUGCAGUAUCCUCCUUUUUCUCUUU
.......((((((((((((((((((...((((((.((........((((((....)))))).((((.(.....).))))..)).)))))))))))))))))))))))). ( -42.80)
>363120_ENSDARG00000014259_ZEBRAFISH_7958_8067/1-114
UCCGUCAUUGGGCCGGGAGGAGGGAAGACCUGGAAGGAGGAAACAACGGUGGAGGGGUUGAACCUACAGACAAUGUGCCUCUCUGUUCCACCGACUCCUCUCUUCCCCUU
((((....))))..((((((((((.((.((.....)).(....)..((((((((.((..((...((((.....))))..)).)).)))))))).)).))))))))))... ( -41.60)
>363120_ENSMUSG00000025932_MOUSE_8201_8311/1-114
UCCUGACAAGGGGAGGGGAGGAGGAUAGACCUGCAGGAAGAAACAACAGUUGAGCCCUCCACUUAAAGACACUAUGCCUUUCCUGGCUGCAGUAUCCUCCCUUUUCUCGUU
((((....))))(((..((((((((((...((((((((.(.(((....)))...)..)))...............(((......)))))))))))))).))))..)))... ( -37.40)
>363120_ENSRNOG00000007590_RAT_8241_8351/1-114
UCUUGACAAGGGGAGGGGAGGAGGAGAGACCUGCAGGAAGAAACAGCAGUUGAGCCCUCCACUUAAAGAUACUAUGCCUUUCCUGGCCGCAGUAUCCUCCCUUUUCUCUUU
.......((((..(((((((((((.(.(((.(((.(.......).))))))...)))))).......((((((..(((......)))...)))))).))))))..).))). ( -37.80)
>consensus
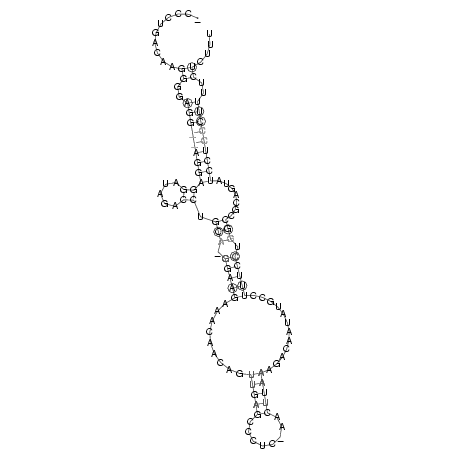
_CCCUGACAAGGGGAGG__AGGAGGAUAGACCUGCA_GGAAGAAACAACAGUUGAGCCCUC_AACUUAAAGACAAUAUGCCUUUCCUGGCCGCAGUAUCCUCCCUUUUCUCUUU
..........((..(((((((((((.....)).(((.(((((.........(((((........)))))............))))).))).......)))))))))..)).... (-19.66 = -20.23 + 0.56)
363120.rnaz
RNAalifold consensus structure
Stefan Washietl
Last modified: Thu Aug 12 14:35:44 CEST 2004