Index >
Results for CNB 359973-rev
Input Alignment
CLUSTAL W(1.81) multiple sequence alignment
359973_ENSG00000121297_HUMAN_9110_9264/1-164 CAAACATAATTTGCCACCTTATTCTTCTCAAGGGGCAACAGCTTGAAATTAAAACTAAAT
359973_ENSMUSG00000043684_MOUSE_8698_8852/1-164 CAAACATAATTTGCCACCTTATTCTTCTCAAGGGGCAACAGCTTGAAATTAAAACTAAAT
359973_SINFRUG00000146762_FUGU_9413_9573/1-164 -AAACATAATGCACCATTTTATTTATCTCAGGGCGCAACAGCTTGAAATGAAAACTAAAT
359973_ENSRNOG00000013938_RAT_9649_9802/1-164 -AAACATAATTTGCCACCTTATTCTTCTCAAGGGGCAACAGCTTGAAATTAAAACTAAAT
********* *** ***** ***** ** *************** **********
359973_ENSG00000121297_HUMAN_9110_9264/1-164 TTGAAATTAATGAAGCACATTCTGGAGGTGGCATTCATTTCTAAAGTAATAAATTACTTG
359973_ENSMUSG00000043684_MOUSE_8698_8852/1-164 TTGAAATTAATGAAGCACATTCTGGAGGTGGCATTCACTTCTAAAGTAATAAATTACTTG
359973_SINFRUG00000146762_FUGU_9413_9573/1-164 CTGAAATTAATGAAGCCCAATCTCACTGTGGCATTCTCCTCTGGGGTAATAAATGAGTTG
359973_ENSRNOG00000013938_RAT_9649_9802/1-164 TTGAAATTAATGAAGCACATTCTGGAGGTGGCATTCACTTCTAAAGTAATAAATTACTTG
*************** ** *** ********* *** ********* * ***
359973_ENSG00000121297_HUMAN_9110_9264/1-164 TATCAACCTCAGAGACAGCAGTTCCTGTCTGTCA---------A
359973_ENSMUSG00000043684_MOUSE_8698_8852/1-164 TATCAACCTCAGAGACAGCAGTTCCTGTCTGTCA---------A
359973_SINFRUG00000146762_FUGU_9413_9573/1-164 GATCAACCTC-CATCTGTCAGTTAATATGTCTGACGCCTCTTC-
359973_ENSRNOG00000013938_RAT_9649_9802/1-164 TATCAACCTCAGAGACAGCAGTTCCTGTCTGTCA---------A
********* * ***** * * * * *
359973-rev.aln
RNAz output
############################ RNAz 0.1 ##############################
Sequences: 4
Columns: 164
Mean pairwise identity: 84.21
Mean single sequence MFE: -34.15
Consensus MFE: -21.67
Energy contribution: -21.67
Covariance contribution: 0.00
Mean z-score: -1.97
Structure conservation index: 0.63
SVM decision value: 0.34
SVM RNA-class probability: 0.694057
Prediction: RNA
######################################################################
>359973_ENSG00000121297_HUMAN_9110_9264/1-164
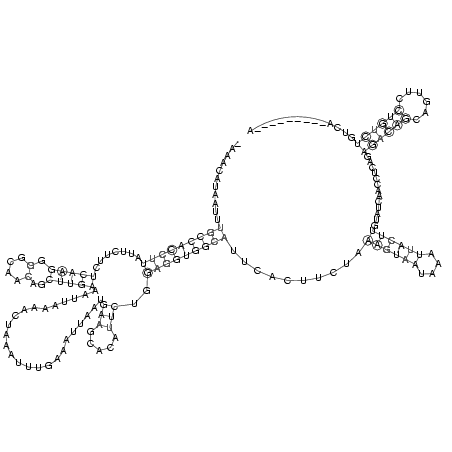
CAAACAUAAUUUGCCACCUUAUUCUUCUCAAGGGGCAACAGCUUGAAAUUAAAACUAAAUUUGAAAUUAAUGAAGCACAUUCUGGAGGUGGCAUUCAUUUCUAAAGUAAUAAAUUACUUGUAUCAACCUCAGAGACAGCAGUUCCUGUCUGUCAA
...........(((((((((...(((((((((.(....)..)))))..(((((......))))).......)))).........)))))))))..........((((((....))))))..............((((((((...))).))))).. ( -34.20)
>359973_ENSMUSG00000043684_MOUSE_8698_8852/1-164
CAAACAUAAUUUGCCACCUUAUUCUUCUCAAGGGGCAACAGCUUGAAAUUAAAACUAAAUUUGAAAUUAAUGAAGCACAUUCUGGAGGUGGCAUUCACUUCUAAAGUAAUAAAUUACUUGUAUCAACCUCAGAGACAGCAGUUCCUGUCUGUCAA
...........(((((((((...(((((((((.(....)..)))))..(((((......))))).......)))).........)))))))))..........((((((....))))))..............((((((((...))).))))).. ( -34.20)
>359973_SINFRUG00000146762_FUGU_9413_9573/1-164
AAACAUAAUGCACCAUUUUAUUUAUCUCAGGGCGCAACAGCUUGAAAUGAAAACUAAAUCUGAAAUUAAUGAAGCCCAAUCUCACUGUGGCAUUCUCCUCUGGGGUAAUAAAUGAGUUGGAUCAACCUCCAUCUGUCAGUUAAUAUGUCUGACGCCUCUUC
.........((..(((((...(((((((((((.((....)).............................((((((((.......)).))).)))..))))))))))).)))))((.((((......)))).))(((((.........)))))))...... ( -34.00)
>359973_ENSRNOG00000013938_RAT_9649_9802/1-164
AAACAUAAUUUGCCACCUUAUUCUUCUCAAGGGGCAACAGCUUGAAAUUAAAACUAAAUUUGAAAUUAAUGAAGCACAUUCUGGAGGUGGCAUUCACUUCUAAAGUAAUAAAUUACUUGUAUCAACCUCAGAGACAGCAGUUCCUGUCUGUCAA
..........(((((((((...(((((((((.(....)..)))))..(((((......))))).......)))).........)))))))))..........((((((....))))))..............((((((((...))).))))).. ( -34.20)
>consensus
_AAACAUAAUUUGCCACCUUAUUCUUCUCAAGGGGCAACAGCUUGAAAUUAAAACUAAAUUUGAAAUUAAUGAAGCACAUUCUGGAGGUGGCAUUCACUUCUAAAGUAAUAAAUUACUUGUAUCAACCUCAGAGACAGCAGUUCCUGUCUGUCA_________A
...........(((((((((.......(((((.(....)..))))).........................(((.....)))..)))))))))..........((((((....)))))).............((((((......)))))).............. (-21.67 = -21.67 + 0.00)
359973-rev.rnaz
RNAalifold consensus structure
Stefan Washietl
Last modified: Thu Aug 12 14:35:44 CEST 2004