Index >
Results for CNB 284325
Input Alignment
CLUSTAL W(1.81) multiple sequence alignment
284325_ENSG00000183386_HUMAN_8149_8251/1-109 G-AAACCTGGGCCTGAGTGGCAGGA-AGGGGGCTGAGAAAGAGGAG--TATCTTC--TCT
284325_ENSMUSG00000032643_MOUSE_8110_8210/1-109 G-AAACCAGGACCTGAATGACAGGG--GAGAGAGAAGGAGGGGCTC--TCTCCAC--CCT
284325_ENSDARG00000011937_ZEBRAFISH_4131_4233/1-109 G-AAACCTGGGCCTGAGTGGCAGGA-AGGGGGCTGAGAAAGAGGAG--TATCTTC--TCT
284325_ENSRNOG00000007541_RAT_8103_8209/1-109 -AAGACCTGAATGACGGTGGGGGGAGAGAAGGAGAGGGAAGAGGAGGTTCTCTTCATCCT
* *** * ** ** * * * * * * * ** * **
284325_ENSG00000183386_HUMAN_8149_8251/1-109 ACACCATAGCCCCCAGCCCAGGAGTTCCAGCTCTAAAGACCCTGGCCCC
284325_ENSMUSG00000032643_MOUSE_8110_8210/1-109 TCAGCATGACCGCTACCTGAGGACTCTCCACTTTGAAGACTCCCACTT-
284325_ENSDARG00000011937_ZEBRAFISH_4131_4233/1-109 ACACCATAGCCCCCAGCCCAGGAGTTCCAGCTCTAAAGACCCTGGCCCC
284325_ENSRNOG00000007541_RAT_8103_8209/1-109 TTAGCATAACCCCTACCTGAGGACTCTCCCCTTTGAAGACTTCAGCTT-
* *** ** * * * **** * * ** * ***** *
//
284325.aln
RNAz output
############################ RNAz 0.1 ##############################
Sequences: 4
Columns: 109
Mean pairwise identity: 65.04
Mean single sequence MFE: -38.91
Consensus MFE: -15.68
Energy contribution: -18.05
Covariance contribution: 2.37
Mean z-score: -2.23
Structure conservation index: 0.40
SVM decision value: 0.56
SVM RNA-class probability: 0.782945
Prediction: RNA
######################################################################
>284325_ENSG00000183386_HUMAN_8149_8251/1-109
GAAACCUGGGCCUGAGUGGCAGGAAGGGGGCUGAGAAAGAGGAGUAUCUUCUCUACACCAUAGCCCCCAGCCCAGGAGUUCCAGCUCUAAAGACCCUGGCCCC
....((((((((((.....)))...((((((((.(..(((((((...)))))))....).)))))))).))))))).(..((((.((....))..))))..). ( -43.30)
>284325_ENSMUSG00000032643_MOUSE_8110_8210/1-109
GAAACCAGGACCUGAAUGACAGGGGAGAGAGAAGGAGGGGCUCUCUCCACCCUUCAGCAUGACCGCUACCUGAGGACUCUCCACUUUGAAGACUCCCACUU
.......(((((((.....))))((((((.(((((..(((.....)))..)))))(((......))).........))))))...........)))..... ( -31.10)
>284325_ENSDARG00000011937_ZEBRAFISH_4131_4233/1-109
GAAACCUGGGCCUGAGUGGCAGGAAGGGGGCUGAGAAAGAGGAGUAUCUUCUCUACACCAUAGCCCCCAGCCCAGGAGUUCCAGCUCUAAAGACCCUGGCCCC
....((((((((((.....)))...((((((((.(..(((((((...)))))))....).)))))))).))))))).(..((((.((....))..))))..). ( -43.30)
>284325_ENSRNOG00000007541_RAT_8103_8209/1-109
AAGACCUGAAUGACGGUGGGGGGAGAGAAGGAGAGGGAAGAGGAGGUUCUCUUCAUCCUUUAGCAUAACCCCUACCUGAGGACUCUCCCCUUUGAAGACUUCAGCUU
.....(((((.......(((((((((((((((....(((((((....))))))).)))))..........(((.....))).)))))))))).......)))))... ( -37.94)
>consensus
G_AAACCUGGACCUGAGUGGCAGGA_AGAGGGAGAAGAAAGAGGAG__UAUCUUC__CCUACACCAUAACCCCCACCCCAGGACUCCCAGCUCUAAAGACCCCGGCCC_
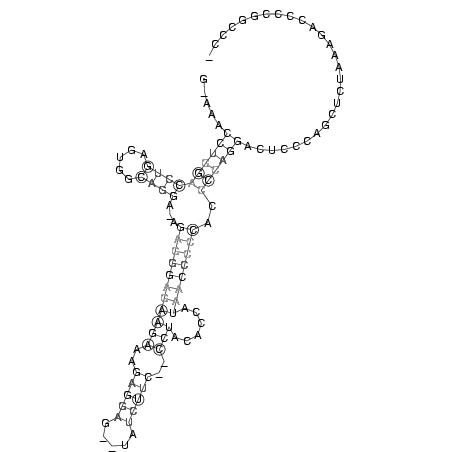
.....((((((((((.....))))...(((((((((((..(((((.....)))))..)))......))))))))..))))))........................... (-15.68 = -18.05 + 2.37)
284325.rnaz
RNAalifold consensus structure
Stefan Washietl
Last modified: Thu Aug 12 14:35:44 CEST 2004