Index >
Results for CNB 264095
Input Alignment
CLUSTAL W(1.81) multiple sequence alignment
264095_ENSG00000185045_HUMAN_8824_8926/1-104 -CCTCAGTTTCACTGCCATGGTTTCCACTCCTGAGAACCACCAGACTTTCATCAACTCAG
264095_ENSMUSG00000043873_MOUSE_5451_5554/1-104 CCCTCAGGTTCAGTGCCATGGTCTCCACTCCACACAACCAACAGACTTTCATTAACTCAG
264095_ENSRNOG00000017574_RAT_8751_8853/1-104 CCCTTAGTTTTATCACCATGGTGTCAACTCCTGAGAGCCGCCACACTTTCATTACTTCAG
264095_SINFRUG00000153198_FUGU_1686_1786/1-104 CC---AGATTCACCACAATGGTGTCAACACAAACCAGCAGAAGCAAATTCATCCAGTCCT
* ** ** * * ***** ** ** * * * * ***** **
264095_ENSG00000185045_HUMAN_8824_8926/1-104 TCATCAAATTCCTGCGCCAGTATGAGTTTGACGGGCTGGACTTT
264095_ENSMUSG00000043873_MOUSE_5451_5554/1-104 CCATCAAATTCCTGCGCCAGTATGGGTTTGATGGACTGAACCTG
264095_ENSRNOG00000017574_RAT_8751_8853/1-104 TTAGAAAATTCCTGCGAAAGTATGGATTTGATGGACTTAACTT-
264095_SINFRUG00000153198_FUGU_1686_1786/1-104 CCATCAAACAGCTGAGAAAATACGGATTTGATGGCCTGGATCTG
* *** *** * * ** * ***** ** ** * *
//
264095.aln
RNAz output
############################ RNAz 0.1 ##############################
Sequences: 4
Columns: 104
Mean pairwise identity: 68.91
Mean single sequence MFE: -25.95
Consensus MFE: -13.93
Energy contribution: -14.86
Covariance contribution: 0.94
Mean z-score: -2.19
Structure conservation index: 0.54
SVM decision value: 2.19
SVM RNA-class probability: 0.989914
Prediction: RNA
######################################################################
>264095_ENSG00000185045_HUMAN_8824_8926/1-104
CCUCAGUUUCACUGCCAUGGUUUCCACUCCUGAGAACCACCAGACUUUCAUCAACUCAGUCAUCAAAUUCCUGCGCCAGUAUGAGUUUGACGGGCUGGACUUU
....((((...(((...(((((..((....))..))))).))).............(((((.((((((((.(((....))).))))))))..))))))))).. ( -26.40)
>264095_ENSMUSG00000043873_MOUSE_5451_5554/1-104
CCCUCAGGUUCAGUGCCAUGGUCUCCACUCCACACAACCAACAGACUUUCAUUAACUCAGCCAUCAAAUUCCUGCGCCAGUAUGGGUUUGAUGGACUGAACCUG
....((((((((((....(((........)))............................(((((((((((.(((....))).))))))))))))))))))))) ( -33.60)
>264095_ENSRNOG00000017574_RAT_8751_8853/1-104
CCCUUAGUUUUAUCACCAUGGUGUCAACUCCUGAGAGCCGCCACACUUUCAUUACUUCAGUUAGAAAAUUCCUGCGAAAGUAUGGAUUUGAUGGACUUAACUU
..(((((......(((....))).......))))).....(((((..(((((.((((..((..((....))..))..)))))))))..)).)))......... ( -19.32)
>264095_SINFRUG00000153198_FUGU_1686_1786/1-104
CCAGAUUCACCACAAUGGUGUCAACACAAACCAGCAGAAGCAAAUUCAUCCAGUCCUCCAUCAAACAGCUGAGAAAAUACGGAUUUGAUGGCCUGGAUCUG
.((((..((((.....)))).............((....)).......(((((....((((((((...(((........))).)))))))).))))))))) ( -24.50)
>consensus
CCCUCAGUUUCACCACCAUGGUGUCAACUCCAGACAACCAACACACUUUCAUCAACUCAGCCAUCAAAUUCCUGCGAAAGUAUGGAUUUGAUGGACUGAACCUG
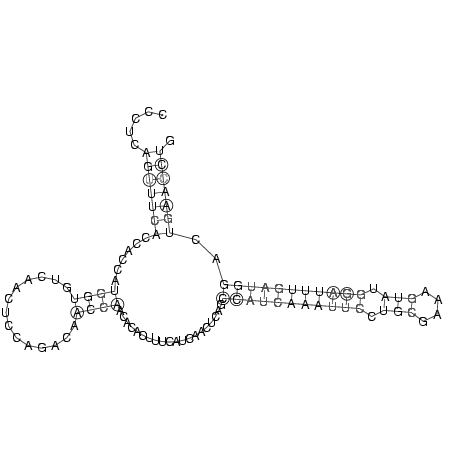
.....(((((((......((((..............))))....................(((((((((((.(((....))).)))))))))))..))))))). (-13.93 = -14.86 + 0.94)
264095.rnaz
RNAalifold consensus structure
Stefan Washietl
Last modified: Thu Aug 12 14:35:44 CEST 2004