Index >
Results for CNB 23410
Input Alignment
CLUSTAL W(1.81) multiple sequence alignment
23410_ENSMUSG00000027022_MOUSE_1204_1246/1-55 -----------GATGTTAGAACCTGCATGTGGCTGTTTGAAACTCAGCCACTTG-
23410_SINFRUG00000130283_FUGU_5276_5326/1-55 ---TGAGCCACGACGTAAGAGAAGCTCGGTGGATGTTTGAAACCAAACCGCTTG-
23410_ENSG00000168334_HUMAN_7381_7433/1-55 -TTTGAGACCCTTCCCTTGGACA-GCATTGGACAGGGTGAGGTTCTGGCCCATGG
23410_ENSDARG00000020243_ZEBRAFISH_7552_7594/1-55 -----------GATGTCCGTAGAAATTGTTGGGTGTTTGAGACACAACCGATGG-
23410_ENSRNOG00000018308_RAT_6563_6616/1-55 CTTCGCACTTCACAGGAAGCAGAAGGCAGCGTCTTCTAGGAACCCAAAGTATGG-
* . . * : :*... .:. .: *
//
23410.aln
RNAz output
############################ RNAz 0.1 ##############################
Sequences: 5
Columns: 55
Mean pairwise identity: 38.93
Mean single sequence MFE: -11.56
Consensus MFE: -1.48
Energy contribution: -1.60
Covariance contribution: 0.12
Mean z-score: -0.33
Structure conservation index: 0.13
SVM decision value: 0.00
SVM RNA-class probability: 0.533916
Prediction: RNA
######################################################################
>23410_ENSMUSG00000027022_MOUSE_1204_1246/1-55
GAUGUUAGAACCUGCAUGUGGCUGUUUGAAACUCAGCCACUUG
.((((........))))(((((((.........)))))))... ( -13.00)
>23410_SINFRUG00000130283_FUGU_5276_5326/1-55
UGAGCCACGACGUAAGAGAAGCUCGGUGGAUGUUUGAAACCAAACCGCUUG
.((((..(..(....).)..))))(((((...((((....))))))))).. ( -11.00)
>23410_ENSG00000168334_HUMAN_7381_7433/1-55
UUUGAGACCCUUCCCUUGGACAGCAUUGGACAGGGUGAGGUUCUGGCCCAUGG
....(((.(((..(((((..(......)..)))))..))).)))......... ( -17.40)
>23410_ENSDARG00000020243_ZEBRAFISH_7552_7594/1-55
GAUGUCCGUAGAAAUUGUUGGGUGUUUGAGACACAACCGAUGG
..............(..(((((((((...)))))..))))..) ( -7.10)
>23410_ENSRNOG00000018308_RAT_6563_6616/1-55
CUUCGCACUUCACAGGAAGCAGAAGGCAGCGUCUUCUAGGAACCCAAAGUAUGG
.(((...((((....)))).(((((((...))))))).)))..(((.....))) ( -9.30)
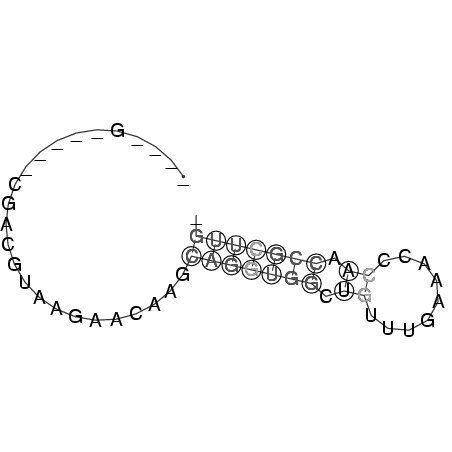
>consensus
____G_____CGACGUAAGAACAAGCAGGUGGCUGUUUGAAACCCAACCGCUUG_
.........................(((((((.((.........)).))))))). ( -1.48 = -1.60 + 0.12)
23410.rnaz
RNAalifold consensus structure
Stefan Washietl
Last modified: Thu Aug 12 14:35:44 CEST 2004