Index >
Results for CNB 211555
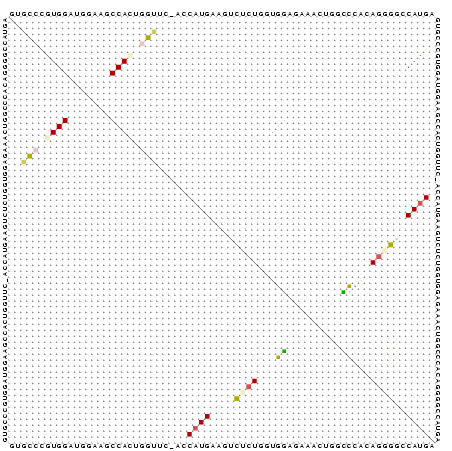
Input Alignment
CLUSTAL W(1.81) multiple sequence alignment
211555_ENSG00000167165_HUMAN_87_157/1-72 GTGCCCATGGATGGGAGCCACTGGTTC-ACCATGAGGTCGGTGGTGGAGAAACTCATTCT
211555_ENSMUSG00000026294_MOUSE_330_401/1-72 CTCTCCATGGATAGAAGCCACACATTTTACCATGAAATCTCTTTTGGATAAACTATCCCA
211555_ENSDARG00000006220_ZEBRAFISH_5196_5265/1-72 ATGCCAGTGGATGGCAGCCATTGGTTG-AGCATGAAGATCCTGGTGGAAGAGATGTCTAG
211555_SINFRUG00000153233_FUGU_2836_2905/1-72 -TGCCTGTGGATGGAAGCCACTGGCTC-AGCATGAAGATACTGGTGAAGGAGCTGGTCCA
211555_ENSRNOG00000018740_RAT_985_1055/1-72 GTGCCTGTGGATAGGAGCCACAAGTGT-ACCATGAAATCTCTTTTGGGTAAACTGGCCCA
* * ***** * ***** * ***** * ** * *
211555_ENSG00000167165_HUMAN_87_157/1-72 CAGGGGGCATGA
211555_ENSMUSG00000026294_MOUSE_330_401/1-72 CAGAGGCCATGA
211555_ENSDARG00000006220_ZEBRAFISH_5196_5265/1-72 TAGGGGTCATG-
211555_SINFRUG00000153233_FUGU_2836_2905/1-72 GAGGGGCCACGA
211555_ENSRNOG00000018740_RAT_985_1055/1-72 CAGAGGCCATGA
** ** ** *
//
211555.aln
RNAz output
############################ RNAz 0.1 ##############################
Sequences: 5
Columns: 72
Mean pairwise identity: 66.81
Mean single sequence MFE: -22.82
Consensus MFE: -6.64
Energy contribution: -6.88
Covariance contribution: 0.24
Mean z-score: -2.25
Structure conservation index: 0.29
SVM decision value: 0.96
SVM RNA-class probability: 0.889915
Prediction: RNA
######################################################################
>211555_ENSG00000167165_HUMAN_87_157/1-72
GUGCCCAUGGAUGGGAGCCACUGGUUCACCAUGAGGUCGGUGGUGGAGAAACUCAUUCUCAGGGGGCAUGA
((((((..(((((((.(((((((....(((....)))))))))).......))))))).....)))))).. ( -27.40)
>211555_ENSMUSG00000026294_MOUSE_330_401/1-72
CUCUCCAUGGAUAGAAGCCACACAUUUUACCAUGAAAUCUCUUUUGGAUAAACUAUCCCACAGAGGCCAUGA
.....(((((.(((((........))))))))))...(((((...(((((...)))))...)))))...... ( -15.60)
>211555_ENSDARG00000006220_ZEBRAFISH_5196_5265/1-72
AUGCCAGUGGAUGGCAGCCAUUGGUUGAGCAUGAAGAUCCUGGUGGAAGAGAUGUCUAGUAGGGGUCAUG
..((((((((.......))))))))....(((((...(((((.((((.......)))).))))).))))) ( -24.70)
>211555_SINFRUG00000153233_FUGU_2836_2905/1-72
UGCCUGUGGAUGGAAGCCACUGGCUCAGCAUGAAGAUACUGGUGAAGGAGCUGGUCCAGAGGGGCCACGA
..(((.(((((((...)))(..((((..(((.(......).)))...))))..))))).)))........ ( -22.90)
>211555_ENSRNOG00000018740_RAT_985_1055/1-72
GUGCCUGUGGAUAGGAGCCACAAGUGUACCAUGAAAUCUCUUUUGGGUAAACUGGCCCACAGAGGCCAUGA
((((((((((.......)))).)).))))((((...(((((..(((((......))))).))))).)))). ( -23.50)
>consensus
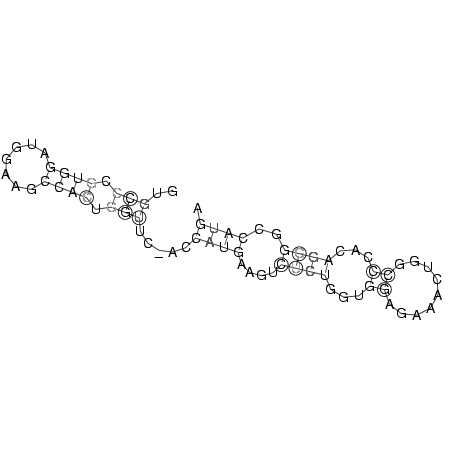
GUGCCCGUGGAUGGAAGCCACUGGUUC_ACCAUGAAGUCUCUGGUGGAGAAACUGGCCCACAGGGGCCAUGA
..(((.((((.......)))).))).....((((....((((...((.........))...))))..)))). ( -6.64 = -6.88 + 0.24)
211555.rnaz
RNAalifold consensus structure
Stefan Washietl
Last modified: Thu Aug 12 14:35:44 CEST 2004