Index >
Results for CNB 160930
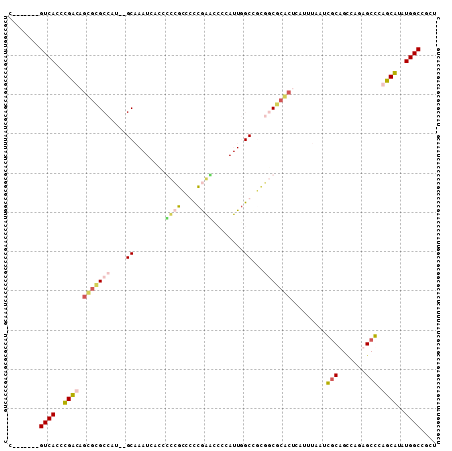
Input Alignment
CLUSTAL W(1.81) multiple sequence alignment
160930_ENSG00000182968_HUMAN_9151_9250/1-109 C-------GTCACTCGCCAGCGCGCCAT--GCAAATCACCGCCGCCGCCGGCTCCCATTG
160930_ENSDARG00000008131_ZEBRAFISH_8806_8907/1-109 C-------GTCACCTGACCGTGAGCCATATGCAAATCAGCCCCGTCCTCAAACGCCATTG
160930_ENSMUSG00000047701_MOUSE_9157_9256/1-109 C-------GTCACTCGCCAGCGCGCCAT--GCAAATCACCGCCGCCGCCGGTTCCCATTG
160930_SINFRUG00000126913_FUGU_9627_9731/1-109 -TCTTTTAGTCACCTGACCAAACGCCAT--GCAAATCATTCTGTTGCTGGAGCCCCATTG
***** * * ***** ******** ******
160930_ENSG00000182968_HUMAN_9151_9250/1-109 GCCGCGGCGCGCTCATTTAATGGCAGCCCGGGCCCGGCGTATGGCTGCT
160930_ENSDARG00000008131_ZEBRAFISH_8806_8907/1-109 GCTCTCTCTCACTCATTTATTCCCTGTCAAAGCGCATCATATGGCCGCT
160930_ENSMUSG00000047701_MOUSE_9157_9256/1-109 GCCGCGGCGCGCTCATTTAATGGCAGCCCGGGCCCGGCGTATGGCTGCT
160930_SINFRUG00000126913_FUGU_9627_9731/1-109 GCCGCTCCAGACTCATTTAACCGCTGTCAGAGCGCATCATATGGCCGC-
** * ******** * * * ** * * ****** **
//
160930.aln
RNAz output
############################ RNAz 0.1 ##############################
Sequences: 4
Columns: 109
Mean pairwise identity: 67.30
Mean single sequence MFE: -33.80
Consensus MFE: -19.75
Energy contribution: -20.57
Covariance contribution: 0.81
Mean z-score: -1.26
Structure conservation index: 0.58
SVM decision value: 0.11
SVM RNA-class probability: 0.587229
Prediction: RNA
######################################################################
>160930_ENSG00000182968_HUMAN_9151_9250/1-109
CGUCACUCGCCAGCGCGCCAUGCAAAUCACCGCCGCCGCCGGCUCCCAUUGGCCGCGGCGCGCUCAUUUAAUGGCAGCCCGGGCCCGGCGUAUGGCUGCU
.((((..((((.....(((((..((((...(((.(((((.((((......))))))))))))...)))).))))).((....))..))))..)))).... ( -43.80)
>160930_ENSDARG00000008131_ZEBRAFISH_8806_8907/1-109
CGUCACCUGACCGUGAGCCAUAUGCAAAUCAGCCCCGUCCUCAAACGCCAUUGGCUCUCUCUCACUCAUUUAUUCCCUGUCAAAGCGCAUCAUAUGGCCGCU
.(((....))).(((.((((((((............((......))(((.(((((.......................))))).).))..))))))))))). ( -21.50)
>160930_ENSMUSG00000047701_MOUSE_9157_9256/1-109
CGUCACUCGCCAGCGCGCCAUGCAAAUCACCGCCGCCGCCGGUUCCCAUUGGCCGCGGCGCGCUCAUUUAAUGGCAGCCCGGGCCCGGCGUAUGGCUGCU
.((((..((((((((((((..((........)).((.((((((....)))))).))))))))))........(((.......))).))))..)))).... ( -43.50)
>160930_SINFRUG00000126913_FUGU_9627_9731/1-109
UCUUUUAGUCACCUGACCAAACGCCAUGCAAAUCAUUCUGUUGCUGGAGCCCCAUUGGCCGCUCCAGACUCAUUUAACCGCUGUCAGAGCGCAUCAUAUGGCCGC
.......(((....))).....((((((..........((...(((((((((....))..)))))))...))......((((.....)))).....))))))... ( -26.40)
>consensus
C_______GUCACCCGACAGCGCGCCAU__GCAAAUCACCCCCGCCCCCGAACCCCAUUGGCCGCGGCGCACUCAUUUAAUCGCAGCCAGAGCCCAGCAUAUGGCCGCU
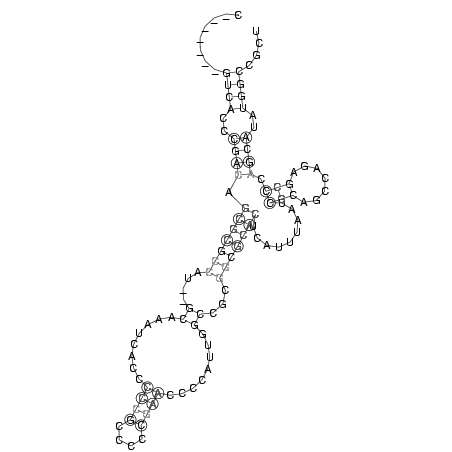
........((((..((((.(((((((....((........((((....))))........))...))))))).........(((.......))).))))..)))).... (-19.75 = -20.57 + 0.81)
160930.rnaz
RNAalifold consensus structure
Stefan Washietl
Last modified: Thu Aug 12 14:35:44 CEST 2004