Index >
Results for CNB 145953
Input Alignment
CLUSTAL W(1.81) multiple sequence alignment
145953_ENSG00000169797_HUMAN_9708_9826/1-125 -CCACCTGCTCCTTGGGGAGC-GCCCATTGTGCCCGCGCCAATTCACAGGCAATTTAGCG
145953_ENSRNOG00000013850_RAT_9660_9778/1-125 -CCACCTGCTCCTCGGGGAGC-GACCATTGTGCCCGCGCCAATTCACAGGCAATTTAGCG
145953_ENSDARG00000009822_ZEBRAFISH_2161_2282/1-125 TCCACCTGCTCCCCAAAGACCAGCCCATTGTTCCCTGAGCAATAAGCACGTCATTTAGCA
145953_ENSMUSG00000048001_MOUSE_9732_9850/1-125 -CCACCTGCTCCTCGGGGAGC-GACCATTGTGCCCGCGCCAATTCACAGGCAATTTAGCG
145953_SINFRUG00000138848_FUGU_3852_3976/1-125 TCCACCTGCTCCCCGGAGACACACTCATTGTCCCCACGGCTATAAACACTTCATTGAGCA
*********** ** ****** *** * ** ** *** ***
145953_ENSG00000169797_HUMAN_9708_9826/1-125 TGCGCTAATGGGCCGGCGCCTTT-GTGCGGCCC-GCCCGGCCATTGGCC-GCGGAGTGTG
145953_ENSRNOG00000013850_RAT_9660_9778/1-125 TGCGCTAATGGGCCGGCGCCTTT-GTGCGGCCG-GCGCCGCCATTGGCC-GCTGAGTGTG
145953_ENSDARG00000009822_ZEBRAFISH_2161_2282/1-125 TGTGGCGGCCCTCTGCTACTTCT-GTCTTTAGA-GCTCTCCCATTGGCT-GCAGACTGTG
145953_ENSMUSG00000048001_MOUSE_9732_9850/1-125 TGCGCTAATGGGCCGGCGCCTTT-GTGCGGCCG-GCGCCGCCATTGGCC-GCCGAGTGTG
145953_SINFRUG00000138848_FUGU_3852_3976/1-125 TGTGGCAGCCCTGCGTTCCTCAAAGCCCTTCCCAGCGTTCCCATTGGCCTGGGGACTGTG
** * * * * ** ******** * ** ****
145953_ENSG00000169797_HUMAN_9708_9826/1-125 GGAA-
145953_ENSRNOG00000013850_RAT_9660_9778/1-125 GGAA-
145953_ENSDARG00000009822_ZEBRAFISH_2161_2282/1-125 AGAAC
145953_ENSMUSG00000048001_MOUSE_9732_9850/1-125 GGAA-
145953_SINFRUG00000138848_FUGU_3852_3976/1-125 AGAAA
***
//
145953.aln
RNAz output
############################ RNAz 0.1 ##############################
Sequences: 5
Columns: 125
Mean pairwise identity: 70.81
Mean single sequence MFE: -49.90
Consensus MFE: -19.34
Energy contribution: -19.14
Covariance contribution: -0.20
Mean z-score: -1.95
Structure conservation index: 0.39
SVM decision value: 0.92
SVM RNA-class probability: 0.881375
Prediction: RNA
######################################################################
>145953_ENSG00000169797_HUMAN_9708_9826/1-125
CCACCUGCUCCUUGGGGAGCGCCCAUUGUGCCCGCGCCAAUUCACAGGCAAUUUAGCGUGCGCUAAUGGGCCGGCGCCUUUGUGCGGCCCGCCCGGCCAUUGGCCGCGGAGUGUGGGAA
......(((((....))))).(((((...((((((((((((...(.(((...(((((....))))).((((((.(((....)))))))))))).)...))))).))))).))))))).. ( -59.40)
>145953_ENSRNOG00000013850_RAT_9660_9778/1-125
CCACCUGCUCCUCGGGGAGCGACCAUUGUGCCCGCGCCAAUUCACAGGCAAUUUAGCGUGCGCUAAUGGGCCGGCGCCUUUGUGCGGCCGGCGCCGCCAUUGGCCGCUGAGUGUGGGAA
(((((((((((....)))))..(((((((((.((((((........)))......))).)))).)))))..(((((((..((.(((((....)))))))..)).))))))).))))... ( -56.40)
>145953_ENSDARG00000009822_ZEBRAFISH_2161_2282/1-125
UCCACCUGCUCCCCAAAGACCAGCCCAUUGUUCCCUGAGCAAUAAGCACGUCAUUUAGCAUGUGGCGGCCCUCUGCUACUUCUGUCUUUAGAGCUCUCCCAUUGGCUGCAGACUGUGAGAAC
(((((((((...((((.(((..((..(((((((...)))))))..))..)))....(((..(((((((....))))))).((((....)))))))......))))..))))...))).)).. ( -33.80)
>145953_ENSMUSG00000048001_MOUSE_9732_9850/1-125
CCACCUGCUCCUCGGGGAGCGACCAUUGUGCCCGCGCCAAUUCACAGGCAAUUUAGCGUGCGCUAAUGGGCCGGCGCCUUUGUGCGGCCGGCGCCGCCAUUGGCCGCCGAGUGUGGGAA
(((((((((((....)))))..(((((((((.((((((........)))......))).)))).)))))..(((((((..((.(((((....)))))))..)).))))))).))))... ( -58.60)
>145953_SINFRUG00000138848_FUGU_3852_3976/1-125
UCCACCUGCUCCCCGGAGACACACUCAUUGUCCCCACGGCUAUAAACACUUCAUUGAGCAUGUGGCAGCCCUGCGUUCCUCAAAGCCCUUCCCAGCGUUCCCAUUGGCCUGGGGACUGUGAGAAA
......(((((....))).))..(((((.(((((((.(((((..(((......(((((.(((..(.....)..)))..))))).((........))))).....)))))))))))).)))))... ( -41.30)
>consensus
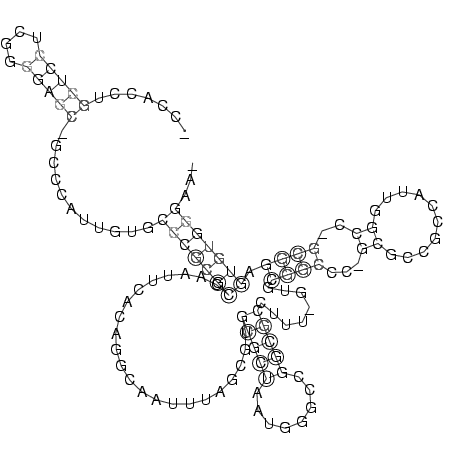
_CCACCUGCUCCUCGGGGAGC_GCCCAUUGUGCCCGCGCCAAUUCACAGGCAAUUUAGCGUGCGCUAAUGGGCCGGCGCCUUU_GUGCGGCCC_GCGCCGCCAUUGGCC_GCGGAGUGUGGGAA_
.......(((((....)))))...........(((((((......................(((((........)))))........((((...((..........))..)))).)))))))... (-19.34 = -19.14 + -0.20)
145953.rnaz
RNAalifold consensus structure
Stefan Washietl
Last modified: Thu Aug 12 14:35:44 CEST 2004