Index >
Results for CNB 143704-rev
Input Alignment
CLUSTAL W(1.81) multiple sequence alignment
143704_ENSG00000165655_HUMAN_9502_9616/1-116 TCTGGCGAGGAAACTCACTTCAAAAGCAGCTGCACCAAGGGGGAGATTAAAGCCTTTGCC
143704_ENSMUSG00000039081_MOUSE_9374_9488/1-116 TCTGGCGAGGAAACTCACTTCAAAAGCAGCTGCACCGAGGGGGAGATTAAAGCCTTTGCC
143704_ENSRNOG00000014237_RAT_9547_9661/1-116 TCTGGCGAGGAAACTCACTTCAAAAGCAGCTGCACGGAGGGGGAGATTAAAGCCTTTGCC
143704_SINFRUG00000138200_FUGU_9523_9635/1-116 -CTGGTAGAAAATTTCACTTCAAAAGCAGCTG-AATTAGTCCGAGATTAAAGCCTTTGCC
**** ** ****************** * ** ******************
143704_ENSG00000165655_HUMAN_9502_9616/1-116 GCCTTCCAATCAAATGGGAGCCCGAGGTGATTGGAGGGTCAAGGGGATGTGACG-C
143704_ENSMUSG00000039081_MOUSE_9374_9488/1-116 GCCTTCCAATCAAATGGGAGCCCGAGGTGATTGGAGGGTCAAGGGGATGTGACG-C
143704_ENSRNOG00000014237_RAT_9547_9661/1-116 GCCTTCCAATCAAATGGGAGCCCGAGGTGATTGGAGGGTCAAGGGGATGTGACG-C
143704_SINFRUG00000138200_FUGU_9523_9635/1-116 GCCTTCCAATCAAATGGGACCCTGAGGTGATTGGAGGGTCAAGGGGATGTGACGT-
******************* ** *******************************
143704-rev.aln
RNAz output
############################ RNAz 0.1 ##############################
Sequences: 4
Columns: 116
Mean pairwise identity: 91.20
Mean single sequence MFE: -39.28
Consensus MFE: -33.95
Energy contribution: -33.33
Covariance contribution: -0.63
Mean z-score: -1.53
Structure conservation index: 0.86
SVM decision value: 0.33
SVM RNA-class probability: 0.689753
Prediction: RNA
######################################################################
>143704_ENSG00000165655_HUMAN_9502_9616/1-116
UCUGGCGAGGAAACUCACUUCAAAAGCAGCUGCACCAAGGGGGAGAUUAAAGCCUUUGCCGCCUUCCAAUCAAAUGGGAGCCCGAGGUGAUUGGAGGGUCAAGGGGAUGUGACGC
....(((.(....)((((.((....((....)).((..((.((((........)))).))((((((((((((..(((....)))...))))))))))))...)).)).))))))) ( -41.20)
>143704_ENSMUSG00000039081_MOUSE_9374_9488/1-116
UCUGGCGAGGAAACUCACUUCAAAAGCAGCUGCACCGAGGGGGAGAUUAAAGCCUUUGCCGCCUUCCAAUCAAAUGGGAGCCCGAGGUGAUUGGAGGGUCAAGGGGAUGUGACGC
....(((.(....)((((.((....((....)).((..((.((((........)))).))((((((((((((..(((....)))...))))))))))))...)).)).))))))) ( -41.20)
>143704_ENSRNOG00000014237_RAT_9547_9661/1-116
UCUGGCGAGGAAACUCACUUCAAAAGCAGCUGCACGGAGGGGGAGAUUAAAGCCUUUGCCGCCUUCCAAUCAAAUGGGAGCCCGAGGUGAUUGGAGGGUCAAGGGGAUGUGACGC
(((...(((....))).((((....((....))...)))).)))........((((((..((((((((((((..(((....)))...)))))))))))))))))).......... ( -41.40)
>143704_SINFRUG00000138200_FUGU_9523_9635/1-116
CUGGUAGAAAAUUUCACUUCAAAAGCAGCUGAAUUAGUCCGAGAUUAAAGCCUUUGCCGCCUUCCAAUCAAAUGGGACCCUGAGGUGAUUGGAGGGUCAAGGGGAUGUGACGU
.............((((.((.............((((((...))))))..((((((..((((((((((((..(.((...)).)..)))))))))))))))))))).))))... ( -33.30)
>consensus
UCUGGCGAGGAAACUCACUUCAAAAGCAGCUGCACCGAGGGGGAGAUUAAAGCCUUUGCCGCCUUCCAAUCAAAUGGGAGCCCGAGGUGAUUGGAGGGUCAAGGGGAUGUGACG_C
...((.(((....))).))......(((.((.(.(....).).)).......((((((..((((((((((((..(((....)))...))))))))))))))))))..)))...... (-33.95 = -33.33 + -0.63)
143704-rev.rnaz
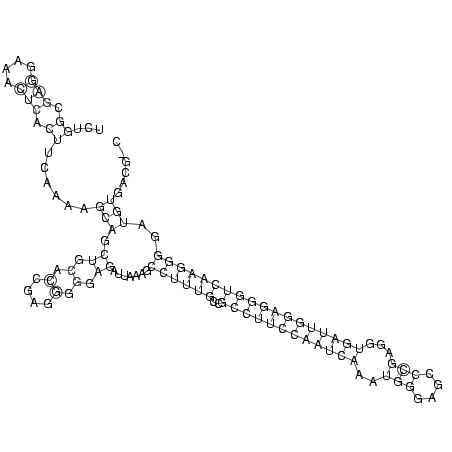
RNAalifold consensus structure
Stefan Washietl
Last modified: Thu Aug 12 14:35:44 CEST 2004