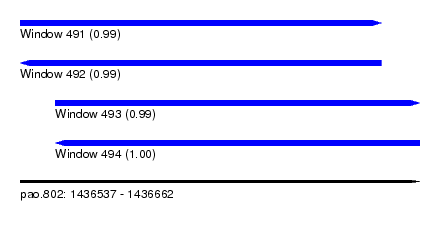
| Sequence ID | pao.802 |
|---|---|
| Location | 1,436,537 – 1,436,662 |
| Length | 125 |
| Max. P | 0.996479 |

| Location | 1,436,537 – 1,436,650 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.48 |
| Mean single sequence MFE | -37.69 |
| Consensus MFE | -27.69 |
| Energy contribution | -27.83 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.73 |
| SVM decision value | 2.38 |
| SVM RNA-class probability | 0.993176 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.802 1436537 113 + 6264404 CCGCCGC-AGCAUUAUUACCGCCAUUCCAGCUA--UGGCGGGUUAGCCAGCGUCC---ACUCCGAACCCGCCCUCGAGGCGGGUUCUU-CGUUCUGUCUCCUGGGCCAAAACCACCGAAC ..(((.(-((........(((((((.......)--))))))..............---.....(((((((((.....)))))))))..-...........)))))).............. ( -38.00) >pst.2260 4311668 98 + 6397126 CGGCCAG-AUUAUUAUUACCGUCAUUCCAGUCA--UGGCGGGUUAGCCGACGUUU---GCAAAAAACCCGCCCUGGAGGCGGGUUUUU----UCUGUCUCCUGGGC-AA----------- ((((..(-((....))).(((((((.......)--))))))....))))..((((---.....))))..((((.(((((((((.....----))))))))).))))-..----------- ( -38.40) >pss.989 4213940 98 - 6093698 CGGCCAG-AUUAUUAUUACCGUCAUUCCAGUCA--UGGCGGGUUAGCCGACGUUU---GCCAAAAACCCGCCCUGGAGGCGGGUUUCU----UCUGUCUCCCGGGC-AA----------- ((((..(-((....))).(((((((.......)--))))))....))))..((((---.....))))..((((.(((((((((.....----))))))))).))))-..----------- ( -38.40) >pel.1698 2767291 101 - 5888780 CCGCCAGCAUUAUUAUUACCGCCAUUAUCGGUU--UGGCGGGCUAGCGCGUACGU---GCACCGAACCCGCCCUGGAGGCGGGUUUUUUCUCUCUGACUCCUGGGC-------------- ...((((...........((((((.........--))))))....((((....))---))...(((((((((.....)))))))))..............))))..-------------- ( -36.70) >pf5.1854 3504933 99 - 7074893 CCAGCCA-AGCAUUAUUACCGCCAUUCC--UUA-UUGGCGGGCUUGCUGACGACU---GCACCCAACCCGCCCUGGAGGCGGGUUUUUUCUCUCUGUCUCCCGGGC-------------- .((((.(-(((.......((((((....--...-.))))))))))))))......---...........((((.(((((((((.........))))))))).))))-------------- ( -39.71) >pfo.1531 2997812 103 + 6438405 CCAGCCA-AGCAUUAUUACCGCCAUUCC--UCAUUUGGCGGGCUAGCUCACGACUGCAGCACCCAACCCGCCCUAGAGGCGGGUUUUUAUUUUCUGUCUCCGGGGU-------------- .......-..........((((((....--.....))))))(((.((........))))).........((((..((((((((.........))))))))..))))-------------- ( -34.90) >consensus CCGCCAG_AGCAUUAUUACCGCCAUUCCAGUCA__UGGCGGGCUAGCCGACGUCU___GCACCAAACCCGCCCUGGAGGCGGGUUUUU_CU_UCUGUCUCCUGGGC______________ ..................((((((...........))))))............................((((.(((((((((.........))))))))).)))).............. (-27.69 = -27.83 + 0.14)

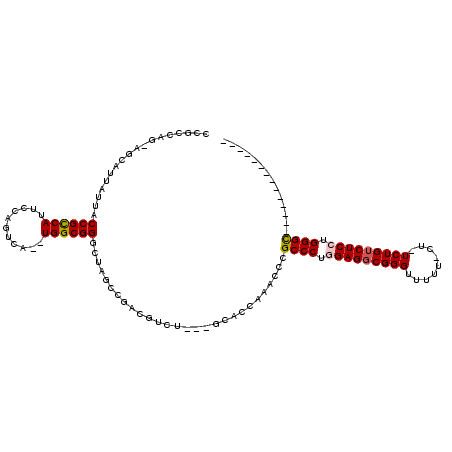
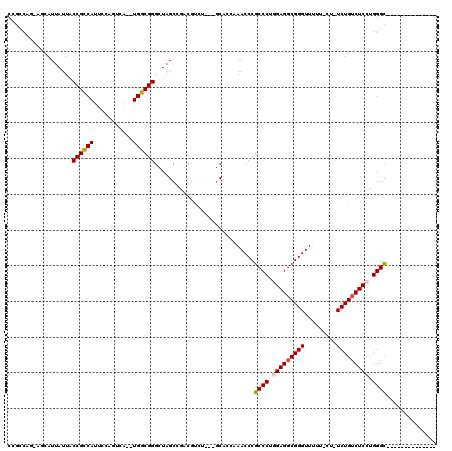
| Location | 1,436,537 – 1,436,650 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.48 |
| Mean single sequence MFE | -40.25 |
| Consensus MFE | -23.64 |
| Energy contribution | -23.95 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.59 |
| SVM decision value | 2.23 |
| SVM RNA-class probability | 0.990782 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.802 1436537 113 - 6264404 GUUCGGUGGUUUUGGCCCAGGAGACAGAACG-AAGAACCCGCCUCGAGGGCGGGUUCGGAGU---GGACGCUGGCUAACCCGCCA--UAGCUGGAAUGGCGGUAAUAAUGCU-GCGGCGG (((.(.((((.((.((((.(....)......-..(((((((((.....))))))))))).))---.)).)))).).)))(((((.--...(......)((((((....))))-))))))) ( -46.80) >pst.2260 4311668 98 - 6397126 -----------UU-GCCCAGGAGACAGA----AAAAACCCGCCUCCAGGGCGGGUUUUUUGC---AAACGUCGGCUAACCCGCCA--UGACUGGAAUGACGGUAAUAAUAAU-CUGGCCG -----------((-((...(....)..(----(((((((((((.....))))))))))))))---))....((((((........--..((((......)))).........-.)))))) ( -35.25) >pss.989 4213940 98 + 6093698 -----------UU-GCCCGGGAGACAGA----AGAAACCCGCCUCCAGGGCGGGUUUUUGGC---AAACGUCGGCUAACCCGCCA--UGACUGGAAUGACGGUAAUAAUAAU-CUGGCCG -----------((-(((..(....)...----(((((((((((.....))))))))))))))---))....((((((........--..((((......)))).........-.)))))) ( -37.45) >pel.1698 2767291 101 + 5888780 --------------GCCCAGGAGUCAGAGAGAAAAAACCCGCCUCCAGGGCGGGUUCGGUGC---ACGUACGCGCUAGCCCGCCA--AACCGAUAAUGGCGGUAAUAAUAAUGCUGGCGG --------------((((.....((.....))...((((((((.....)))))))).)).))---.......(((((((((((((--.........))))))..........))))))). ( -38.40) >pf5.1854 3504933 99 + 7074893 --------------GCCCGGGAGACAGAGAGAAAAAACCCGCCUCCAGGGCGGGUUGGGUGC---AGUCGUCAGCAAGCCCGCCAA-UAA--GGAAUGGCGGUAAUAAUGCU-UGGCUGG --------------((((.(....)..........((((((((.....)))))))))))).(---(((((..((((...((((((.-...--....))))))......))))-)))))). ( -42.90) >pfo.1531 2997812 103 - 6438405 --------------ACCCCGGAGACAGAAAAUAAAAACCCGCCUCUAGGGCGGGUUGGGUGCUGCAGUCGUGAGCUAGCCCGCCAAAUGA--GGAAUGGCGGUAAUAAUGCU-UGGCUGG --------------((((.(....)..........((((((((.....))))))))))))....(((((..((((....((((((.....--....)))))).......)))-)))))). ( -40.70) >consensus ______________GCCCAGGAGACAGA_AG_AAAAACCCGCCUCCAGGGCGGGUUCGGUGC___AGACGUCGGCUAACCCGCCA__UAACUGGAAUGGCGGUAAUAAUAAU_CUGGCGG ..............(((..(....)..........((((((((.....)))))))).......................((((((...........)))))).............))).. (-23.64 = -23.95 + 0.31)
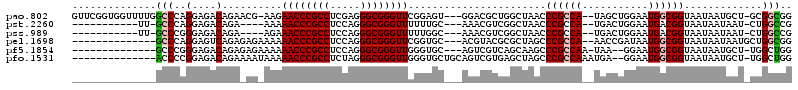
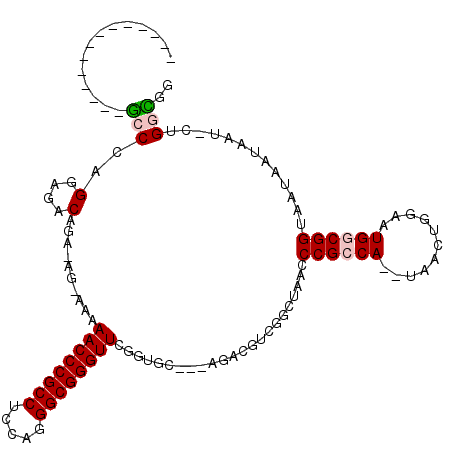
| Location | 1,436,548 – 1,436,662 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.07 |
| Mean single sequence MFE | -36.12 |
| Consensus MFE | -27.69 |
| Energy contribution | -27.83 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.77 |
| SVM decision value | 2.29 |
| SVM RNA-class probability | 0.991760 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
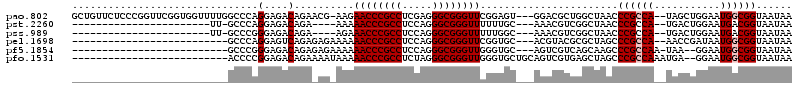
>pao.802 1436548 114 + 6264404 UUAUUACCGCCAUUCCAGCUA--UGGCGGGUUAGCCAGCGUCC---ACUCCGAACCCGCCCUCGAGGCGGGUUCUU-CGUUCUGUCUCCUGGGCCAAAACCACCGAACCGGGAGAACAGC ......(((((((.......)--))))))..............---.....(((((((((.....)))))))))..-....(((((((((((..(.........)..))))))).)))). ( -46.40) >pst.2260 4311679 87 + 6397126 UUAUUACCGUCAUUCCAGUCA--UGGCGGGUUAGCCGACGUUU---GCAAAAAACCCGCCCUGGAGGCGGGUUUUU----UCUGUCUCCUGGGC-AA----------------------- ......(((((((.......)--))))))((.(((....))).---)).........((((.(((((((((.....----))))))))).))))-..----------------------- ( -33.70) >pss.989 4213951 87 - 6093698 UUAUUACCGUCAUUCCAGUCA--UGGCGGGUUAGCCGACGUUU---GCCAAAAACCCGCCCUGGAGGCGGGUUUCU----UCUGUCUCCCGGGC-AA----------------------- ......(((((((.......)--))))))..........((((---.....))))..((((.(((((((((.....----))))))))).))))-..----------------------- ( -33.70) >pel.1698 2767303 89 - 5888780 UUAUUACCGCCAUUAUCGGUU--UGGCGGGCUAGCGCGUACGU---GCACCGAACCCGCCCUGGAGGCGGGUUUUUUCUCUCUGACUCCUGGGC-------------------------- ......((((((.........--))))))(((.((((....))---))...(((((((((.....))))))))).................)))-------------------------- ( -33.50) >pf5.1854 3504944 88 - 7074893 UUAUUACCGCCAUUCC--UUA-UUGGCGGGCUUGCUGACGACU---GCACCCAACCCGCCCUGGAGGCGGGUUUUUUCUCUCUGUCUCCCGGGC-------------------------- ......((((((....--...-.))))))...(((........---)))........((((.(((((((((.........))))))))).))))-------------------------- ( -34.50) >pfo.1531 2997823 92 + 6438405 UUAUUACCGCCAUUCC--UCAUUUGGCGGGCUAGCUCACGACUGCAGCACCCAACCCGCCCUAGAGGCGGGUUUUUAUUUUCUGUCUCCGGGGU-------------------------- ......((((((....--.....))))))(((.((........))))).........((((..((((((((.........))))))))..))))-------------------------- ( -34.90) >consensus UUAUUACCGCCAUUCCAGUCA__UGGCGGGCUAGCCGACGUCU___GCACCAAACCCGCCCUGGAGGCGGGUUUUU_CU_UCUGUCUCCUGGGC__________________________ ......((((((...........))))))............................((((.(((((((((.........))))))))).)))).......................... (-27.69 = -27.83 + 0.14)
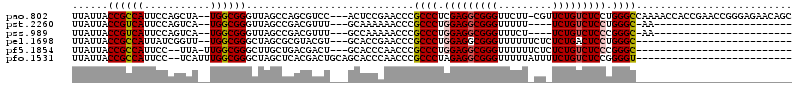
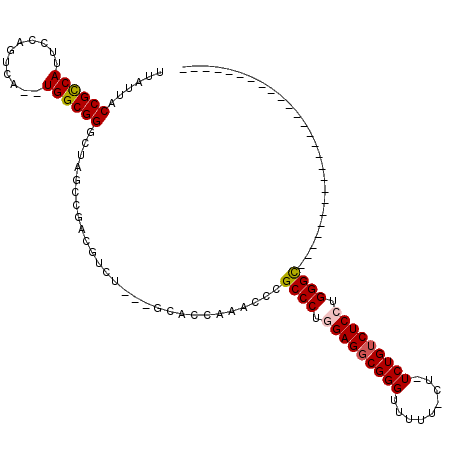
| Location | 1,436,548 – 1,436,662 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.07 |
| Mean single sequence MFE | -36.55 |
| Consensus MFE | -22.40 |
| Energy contribution | -22.73 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.61 |
| SVM decision value | 2.70 |
| SVM RNA-class probability | 0.996479 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.802 1436548 114 - 6264404 GCUGUUCUCCCGGUUCGGUGGUUUUGGCCCAGGAGACAGAACG-AAGAACCCGCCUCGAGGGCGGGUUCGGAGU---GGACGCUGGCUAACCCGCCA--UAGCUGGAAUGGCGGUAAUAA (((((..(.((((((.(((((..((((((..(....)...((.-..(((((((((.....)))))))))...))---.......)))))).))))).--.)))))).)..)))))..... ( -50.20) >pst.2260 4311679 87 - 6397126 -----------------------UU-GCCCAGGAGACAGA----AAAAACCCGCCUCCAGGGCGGGUUUUUUGC---AAACGUCGGCUAACCCGCCA--UGACUGGAAUGACGGUAAUAA -----------------------((-((...(....)..(----(((((((((((.....))))))))))))))---)).(((.(((......))))--))((((......))))..... ( -32.50) >pss.989 4213951 87 + 6093698 -----------------------UU-GCCCGGGAGACAGA----AGAAACCCGCCUCCAGGGCGGGUUUUUGGC---AAACGUCGGCUAACCCGCCA--UGACUGGAAUGACGGUAAUAA -----------------------((-(((..(....)...----(((((((((((.....))))))))))))))---)).(((.(((......))))--))((((......))))..... ( -34.70) >pel.1698 2767303 89 + 5888780 --------------------------GCCCAGGAGUCAGAGAGAAAAAACCCGCCUCCAGGGCGGGUUCGGUGC---ACGUACGCGCUAGCCCGCCA--AACCGAUAAUGGCGGUAAUAA --------------------------((((.((((.(.(.(........)).).)))).))))(((((.(((((---......))))))))))((((--.........))))........ ( -31.70) >pf5.1854 3504944 88 + 7074893 --------------------------GCCCGGGAGACAGAGAGAAAAAACCCGCCUCCAGGGCGGGUUGGGUGC---AGUCGUCAGCAAGCCCGCCAA-UAA--GGAAUGGCGGUAAUAA --------------------------((((.(....)..........((((((((.....))))))))))))((---........))....((((((.-...--....))))))...... ( -34.50) >pfo.1531 2997823 92 - 6438405 --------------------------ACCCCGGAGACAGAAAAUAAAAACCCGCCUCUAGGGCGGGUUGGGUGCUGCAGUCGUGAGCUAGCCCGCCAAAUGA--GGAAUGGCGGUAAUAA --------------------------((((.(....)..........((((((((.....))))))))))))(((((........)).)))((((((.....--....))))))...... ( -35.70) >consensus __________________________GCCCAGGAGACAGA_AG_AAAAACCCGCCUCCAGGGCGGGUUCGGUGC___AGACGUCGGCUAACCCGCCA__UAACUGGAAUGGCGGUAAUAA ...............................(....)..........((((((((.....)))))))).......................((((((...........))))))...... (-22.40 = -22.73 + 0.33)
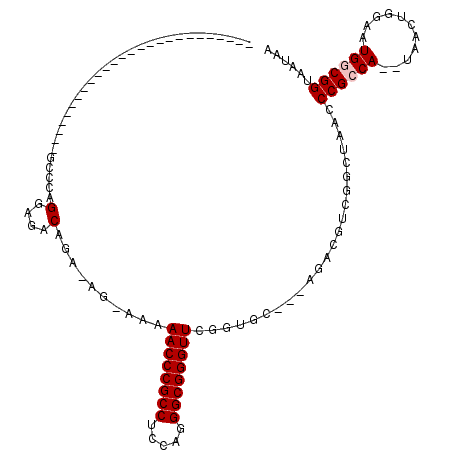
Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:12:15 2007