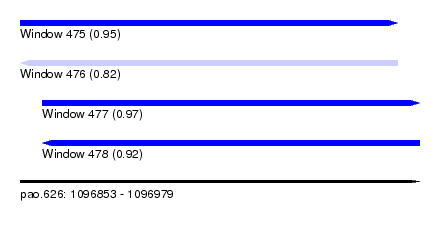
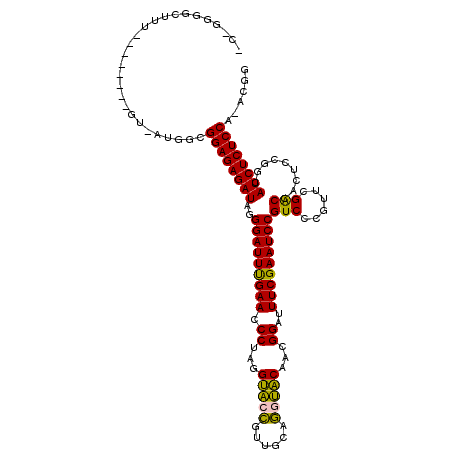
| Sequence ID | pao.626 |
|---|---|
| Location | 1,096,853 – 1,096,979 |
| Length | 126 |
| Max. P | 0.970304 |

| Location | 1,096,853 – 1,096,972 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 83.54 |
| Mean single sequence MFE | -41.85 |
| Consensus MFE | -37.67 |
| Energy contribution | -36.07 |
| Covariance contribution | -1.60 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.947008 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
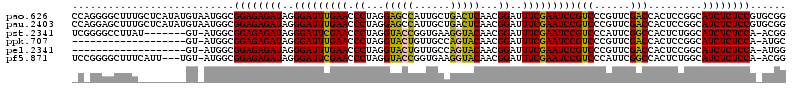
>pao.626 1096853 119 + 6264404 AUGCGCGCCGCACGGAGAGAUGCCGGAGUGGUCGAACGGGACGGAUUCGAAAUCCGUUGAGUCAGCAAUGGCUCCUAGGGUUCAAAUCCCUAUCUCUCCGCCAUUACAUAUGAGCAAAG .((((...)))).((((((((...(((((.((.(.((..(((((((.....)))))))..))...).)).))))).((((.......))))))))))))(((((.....))).)).... ( -43.50) >pau.2403 1984571 119 - 6537648 AUGCGCGCCGCACGGAGAGAUGCCGGAGUGGUCGAACGGGACGGAUUCGAAAUCCGUUGAGUCAGCAAUGGCUCCUAGGGUUCAAAUCCCUAUCUCUCCGCCAUUACAUAUGAGCAAAG .((((...)))).((((((((...(((((.((.(.((..(((((((.....)))))))..))...).)).))))).((((.......))))))))))))(((((.....))).)).... ( -43.50) >pst.2341 1938406 110 - 6397126 AUUCGCGCCGU-UGGAGAGAUGCCAGAGUGGCCGAAUGGGACGGAUUCGAAAUCCGUUGUACCUUCACCGGUACCUAGGGUUCGAAUCCCUAUCUCUCCGCCAU-AC-------AUAAG .........(.-((((((((((((.....)))..........(((((((((.(((...(((((......)))))...))))))))))))..))))))))).)..-..-------..... ( -41.00) >ppk.707 1418329 105 + 6181863 AUGCGCGGCAU-UGGAGAGAUGCCGGAGUGGUCGAACGGGACGGAUUCGAAAUCCGUUGUACUGGCAACAGUACCUAGGGUUCAAAUCCCUAUCUCUCCGCCAU-AC------------ (((.(((((((-(.....))))))((((.((((......)))(((((.(((.(((...((((((....))))))...)))))).)))))....).)))))))))-..------------ ( -43.20) >pel.2341 1531497 105 - 5888780 AUGCGCGCCAU-UGGAGAGAUGCCGGAGUGGUCGAACGGGACGGAUUCGAAAUCCGUUGUACUGGCAACAGUACCUAGGGUUCAAAUCCCUAUCUCUCCGCCAU-AC------------ (((.((.....-.((((((((((((...........)))...(((((.(((.(((...((((((....))))))...)))))).))))).))))))))))))))-..------------ ( -38.90) >pf5.871 1640050 114 + 7074893 AUGCGCGCCGU-UGGAGAGAUGCCAGAGUGGCCGAAUGGGACGGAUUCGAAAUCCGUUGUACCUUCACCGGUACCUAGGGUUCGAAUCCCUAUCUCUCCGCCAU-ACA---AAUGAAAG .........(.-((((((((((((.....)))..........(((((((((.(((...(((((......)))))...))))))))))))..))))))))).)..-...---........ ( -41.00) >consensus AUGCGCGCCGU_UGGAGAGAUGCCGGAGUGGUCGAACGGGACGGAUUCGAAAUCCGUUGUACCAGCAACGGUACCUAGGGUUCAAAUCCCUAUCUCUCCGCCAU_AC________AAAG ............(((((((((((((..(....)...)))...(((((.(((.(((...((((((....))))))...)))))).))))).))))))))))................... (-37.67 = -36.07 + -1.60)

| Location | 1,096,853 – 1,096,972 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 83.54 |
| Mean single sequence MFE | -39.88 |
| Consensus MFE | -35.07 |
| Energy contribution | -33.63 |
| Covariance contribution | -1.44 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.824498 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.626 1096853 119 - 6264404 CUUUGCUCAUAUGUAAUGGCGGAGAGAUAGGGAUUUGAACCCUAGGAGCCAUUGCUGACUCAACGGAUUUCGAAUCCGUCCCGUUCGACCACUCCGGCAUCUCUCCGUGCGGCGCGCAU ..((((......))))..((((((((((((((.......)))))((((...(((..(((...((((((.....))))))...))))))...))))....)))))))))(((...))).. ( -38.20) >pau.2403 1984571 119 + 6537648 CUUUGCUCAUAUGUAAUGGCGGAGAGAUAGGGAUUUGAACCCUAGGAGCCAUUGCUGACUCAACGGAUUUCGAAUCCGUCCCGUUCGACCACUCCGGCAUCUCUCCGUGCGGCGCGCAU ..((((......))))..((((((((((((((.......)))))((((...(((..(((...((((((.....))))))...))))))...))))....)))))))))(((...))).. ( -38.20) >pst.2341 1938406 110 + 6397126 CUUAU-------GU-AUGGCGGAGAGAUAGGGAUUCGAACCCUAGGUACCGGUGAAGGUACAACGGAUUUCGAAUCCGUCCCAUUCGGCCACUCUGGCAUCUCUCCA-ACGGCGCGAAU ....(-------((-.((..((((((((..(((((((((.((...(((((......)))))...))..)))))))))..........(((.....))))))))))).-.))..)))... ( -42.70) >ppk.707 1418329 105 - 6181863 ------------GU-AUGGCGGAGAGAUAGGGAUUUGAACCCUAGGUACUGUUGCCAGUACAACGGAUUUCGAAUCCGUCCCGUUCGACCACUCCGGCAUCUCUCCA-AUGCCGCGCAU ------------(.-.((((((((((((..(((((((((.((...((((((....))))))...))..)))))))))(((......))).........)))))))).-..))))..).. ( -40.90) >pel.2341 1531497 105 + 5888780 ------------GU-AUGGCGGAGAGAUAGGGAUUUGAACCCUAGGUACUGUUGCCAGUACAACGGAUUUCGAAUCCGUCCCGUUCGACCACUCCGGCAUCUCUCCA-AUGGCGCGCAU ------------(.-.((.(((((((((..(((((((((.((...((((((....))))))...))..)))))))))(((......))).........)))))))).-..).))..).. ( -36.30) >pf5.871 1640050 114 - 7074893 CUUUCAUU---UGU-AUGGCGGAGAGAUAGGGAUUCGAACCCUAGGUACCGGUGAAGGUACAACGGAUUUCGAAUCCGUCCCAUUCGGCCACUCUGGCAUCUCUCCA-ACGGCGCGCAU ........---((.-.((.(((((((((..(((((((((.((...(((((......)))))...))..)))))))))..........(((.....))))))))))).-..).))..)). ( -43.00) >consensus CUUU________GU_AUGGCGGAGAGAUAGGGAUUUGAACCCUAGGUACCGUUGCAGGUACAACGGAUUUCGAAUCCGUCCCGUUCGACCACUCCGGCAUCUCUCCA_ACGGCGCGCAU ....................((((((((..(((((((((.((...(((((......)))))...))..)))))))))(((......))).........))))))))............. (-35.07 = -33.63 + -1.44)

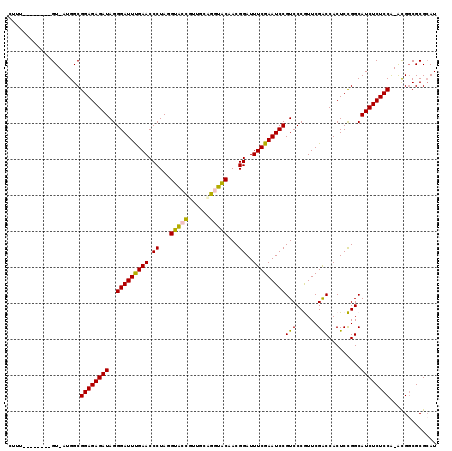
| Location | 1,096,860 – 1,096,979 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 79.66 |
| Mean single sequence MFE | -41.77 |
| Consensus MFE | -37.67 |
| Energy contribution | -36.07 |
| Covariance contribution | -1.60 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970304 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.626 1096860 119 + 6264404 CCGCACGGAGAGAUGCCGGAGUGGUCGAACGGGACGGAUUCGAAAUCCGUUGAGUCAGCAAUGGCUCCUAGGGUUCAAAUCCCUAUCUCUCCGCCAUUACAUAUGAGCAAAGCCCCUGG ..((..((((((((...(((((.((.(.((..(((((((.....)))))))..))...).)).))))).((((.......))))))))))))(((((.....))).))...))...... ( -42.80) >pau.2403 1984578 119 - 6537648 CCGCACGGAGAGAUGCCGGAGUGGUCGAACGGGACGGAUUCGAAAUCCGUUGAGUCAGCAAUGGCUCCUAGGGUUCAAAUCCCUAUCUCUCCGCCAUUACAUAUGAGCAAAGCUCCUGG ..((..((((((((...(((((.((.(.((..(((((((.....)))))))..))...).)).))))).((((.......))))))))))))(((((.....))).))...))...... ( -43.20) >pst.2341 1938413 110 - 6397126 CCGU-UGGAGAGAUGCCAGAGUGGCCGAAUGGGACGGAUUCGAAAUCCGUUGUACCUUCACCGGUACCUAGGGUUCGAAUCCCUAUCUCUCCGCCAU-AC-------AUAAGGCCCCGA .((.-.(((((((((((.....)))..........(((((((((.(((...(((((......)))))...))))))))))))..))))))))(((..-..-------....)))..)). ( -44.20) >ppk.707 1418336 98 + 6181863 GCAU-UGGAGAGAUGCCGGAGUGGUCGAACGGGACGGAUUCGAAAUCCGUUGUACUGGCAACAGUACCUAGGGUUCAAAUCCCUAUCUCUCCGCCAU-AC------------------- ....-.((((((((((((...........)))...(((((.(((.(((...((((((....))))))...)))))).))))).))))))))).....-..------------------- ( -38.70) >pel.2341 1531504 98 - 5888780 CCAU-UGGAGAGAUGCCGGAGUGGUCGAACGGGACGGAUUCGAAAUCCGUUGUACUGGCAACAGUACCUAGGGUUCAAAUCCCUAUCUCUCCGCCAU-AC------------------- ....-.((((((((((((...........)))...(((((.(((.(((...((((((....))))))...)))))).))))).))))))))).....-..------------------- ( -38.70) >pf5.871 1640057 114 + 7074893 CCGU-UGGAGAGAUGCCAGAGUGGCCGAAUGGGACGGAUUCGAAAUCCGUUGUACCUUCACCGGUACCUAGGGUUCGAAUCCCUAUCUCUCCGCCAU-ACA---AAUGAAAGCCCCGGA (((.-.(((((((((((.....)))..........(((((((((.(((...(((((......)))))...))))))))))))..))))))))..(((-...---.))).......))). ( -43.00) >consensus CCGU_UGGAGAGAUGCCGGAGUGGUCGAACGGGACGGAUUCGAAAUCCGUUGUACCAGCAACGGUACCUAGGGUUCAAAUCCCUAUCUCUCCGCCAU_AC________AAAGCCCC_G_ .....(((((((((((((..(....)...)))...(((((.(((.(((...((((((....))))))...)))))).))))).)))))))))).......................... (-37.67 = -36.07 + -1.60)

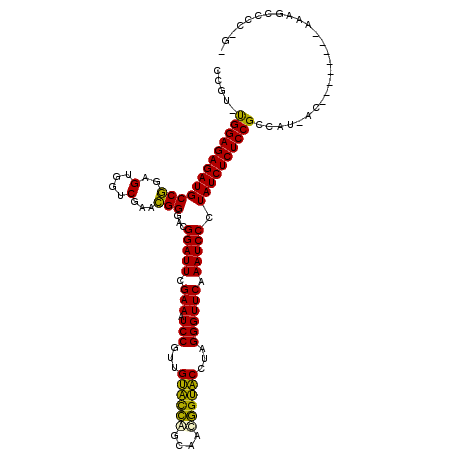
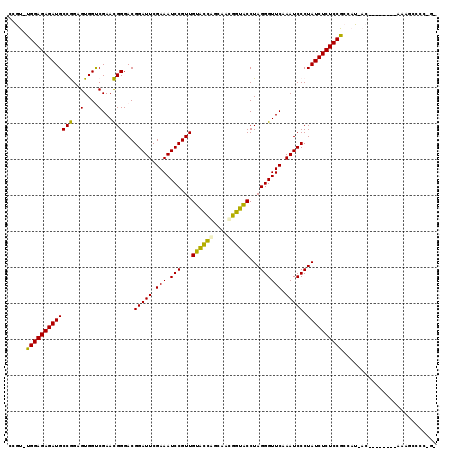
| Location | 1,096,860 – 1,096,979 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 79.66 |
| Mean single sequence MFE | -40.23 |
| Consensus MFE | -35.07 |
| Energy contribution | -33.63 |
| Covariance contribution | -1.44 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922542 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.626 1096860 119 - 6264404 CCAGGGGCUUUGCUCAUAUGUAAUGGCGGAGAGAUAGGGAUUUGAACCCUAGGAGCCAUUGCUGACUCAACGGAUUUCGAAUCCGUCCCGUUCGACCACUCCGGCAUCUCUCCGUGCGG ((..((((...))))..........((((((((((((((.......)))))((((...(((..(((...((((((.....))))))...))))))...))))....)))))))))..)) ( -38.80) >pau.2403 1984578 119 + 6537648 CCAGGAGCUUUGCUCAUAUGUAAUGGCGGAGAGAUAGGGAUUUGAACCCUAGGAGCCAUUGCUGACUCAACGGAUUUCGAAUCCGUCCCGUUCGACCACUCCGGCAUCUCUCCGUGCGG ((..((((...))))..........((((((((((((((.......)))))((((...(((..(((...((((((.....))))))...))))))...))))....)))))))))..)) ( -39.70) >pst.2341 1938413 110 + 6397126 UCGGGGCCUUAU-------GU-AUGGCGGAGAGAUAGGGAUUCGAACCCUAGGUACCGGUGAAGGUACAACGGAUUUCGAAUCCGUCCCAUUCGGCCACUCUGGCAUCUCUCCA-ACGG .((..(((....-------..-..)))((((((((..(((((((((.((...(((((......)))))...))..)))))))))..........(((.....))))))))))).-.)). ( -46.80) >ppk.707 1418336 98 - 6181863 -------------------GU-AUGGCGGAGAGAUAGGGAUUUGAACCCUAGGUACUGUUGCCAGUACAACGGAUUUCGAAUCCGUCCCGUUCGACCACUCCGGCAUCUCUCCA-AUGC -------------------((-((...((((((((..(((((((((.((...((((((....))))))...))..)))))))))(((......))).........)))))))).-)))) ( -35.30) >pel.2341 1531504 98 + 5888780 -------------------GU-AUGGCGGAGAGAUAGGGAUUUGAACCCUAGGUACUGUUGCCAGUACAACGGAUUUCGAAUCCGUCCCGUUCGACCACUCCGGCAUCUCUCCA-AUGG -------------------..-.....((((((((..(((((((((.((...((((((....))))))...))..)))))))))(((......))).........)))))))).-.... ( -35.00) >pf5.871 1640057 114 - 7074893 UCCGGGGCUUUCAUU---UGU-AUGGCGGAGAGAUAGGGAUUCGAACCCUAGGUACCGGUGAAGGUACAACGGAUUUCGAAUCCGUCCCAUUCGGCCACUCUGGCAUCUCUCCA-ACGG .(((..(((..(...---.).-..)))((((((((..(((((((((.((...(((((......)))))...))..)))))))))..........(((.....))))))))))).-.))) ( -45.80) >consensus _C_GGGGCUUU________GU_AUGGCGGAGAGAUAGGGAUUUGAACCCUAGGUACCGUUGCAGGUACAACGGAUUUCGAAUCCGUCCCGUUCGACCACUCCGGCAUCUCUCCA_ACGG ...........................((((((((..(((((((((.((...(((((......)))))...))..)))))))))(((......))).........))))))))...... (-35.07 = -33.63 + -1.44)
Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:11:59 2007