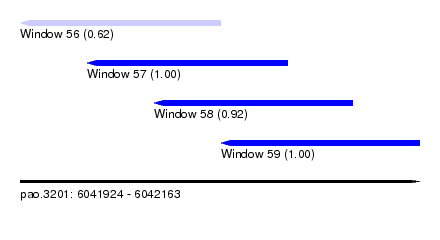
| Sequence ID | pao.3201 |
|---|---|
| Location | 6,041,924 – 6,042,163 |
| Length | 239 |
| Max. P | 0.999716 |

| Location | 6,041,924 – 6,042,044 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.78 |
| Mean single sequence MFE | -37.50 |
| Consensus MFE | -33.38 |
| Energy contribution | -32.77 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.623057 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
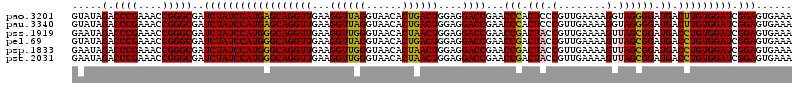
>pao.3201 6041924 120 - 6264404 GAAGGUUAGGUAACACUGACUGGAGGACCGAACCCACUCCCGUUGAAAAGGUAGGGGAUGACUUGUGGAUCGGAGUGAAAGGCUAAUCAAGCUCGGAGAUAGCUGGUUCUCCUCGAAAGC ...((((((......))))))((((((((.....((((((.(((.(.(((...........))).).))).))))))...(((((.((.......))..))))))))))))).(....). ( -38.10) >pau.3340 6314843 120 - 6537648 GAAGGUUAGGUAACACUGACUGGAGGACCGAACCCACUCCCGUUGAAAAGGUAGGGGAUGACUUGUGGAUCGGAGUGAAAGGCUAAUCAAGCUCGGAGAUAGCUGGUUCUCCUCGAAAGC ...((((((......))))))((((((((.....((((((.(((.(.(((...........))).).))).))))))...(((((.((.......))..))))))))))))).(....). ( -38.10) >pss.1919 3849792 120 - 6093698 GAAGGUUGGGUAACACUAACUGGAGGACCGAACCGACUACCGUUGAAAAGUUAGCGGAUGACCUGUGGAUCGGAGUGAAAGGCUAAUCAAGCUCGGAGAUAGCUGGUUCUCCUCGAAAGC ...((((((......))))))((((((((...((((((((.(((((....)))))((....)).)))).)))).......(((((.((.......))..))))))))))))).(....). ( -37.10) >psp.1833 3633796 120 - 5928787 GAAGGUUGGGUAACACUAACUGGAGGACCGAACCGACUACCGUUGAAAAGUUAGCGGAUGACCUGUGGAUCGGAGUGAAAGGCUAAUCAAGCUCGGAGAUAGCUGGUUCUCCUCGAAAGC ...((((((......))))))((((((((...((((((((.(((((....)))))((....)).)))).)))).......(((((.((.......))..))))))))))))).(....). ( -37.10) >pfo.3002 5728701 120 - 6438405 GAAGGUUAGGUAACACUGACUGGAGGACCGAACCGACUACCGUUGAAAAGUUAGCGGAUGACCUGUGGAUCGGAGUGAAAGGCUAAUCAAGCUCGGAGAUAGCUGGUUCUCCUCGAAAGC ...((((((......))))))((((((((...((((((((.(((((....)))))((....)).)))).)))).......(((((.((.......))..))))))))))))).(....). ( -37.50) >pst.2031 3871793 120 - 6397126 GAAGGUUGGGUAACACUAACUGGAGGACCGAACCGACUACCGUUGAAAAGUUAGCGGAUGACCUGUGGAUCGGAGUGAAAGGCUAAUCAAGCUCGGAGAUAGCUGGUUCUCCUCGAAAGC ...((((((......))))))((((((((...((((((((.(((((....)))))((....)).)))).)))).......(((((.((.......))..))))))))))))).(....). ( -37.10) >consensus GAAGGUUAGGUAACACUAACUGGAGGACCGAACCGACUACCGUUGAAAAGUUAGCGGAUGACCUGUGGAUCGGAGUGAAAGGCUAAUCAAGCUCGGAGAUAGCUGGUUCUCCUCGAAAGC ...((((((......))))))(((((((((..(((.(((.(........).)))))).......((...((.((((..............)))).))....))))))))))).(....). (-33.38 = -32.77 + -0.61)
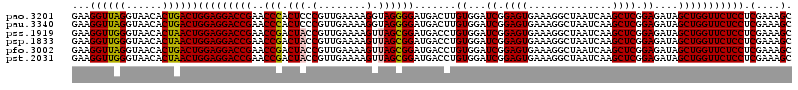
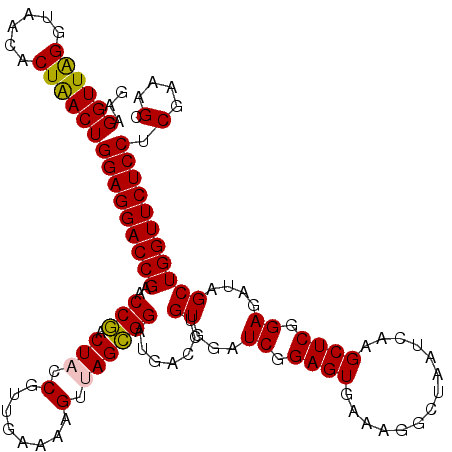
| Location | 6,041,964 – 6,042,084 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -40.17 |
| Consensus MFE | -39.02 |
| Energy contribution | -37.97 |
| Covariance contribution | -1.05 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.97 |
| SVM decision value | 3.94 |
| SVM RNA-class probability | 0.999716 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
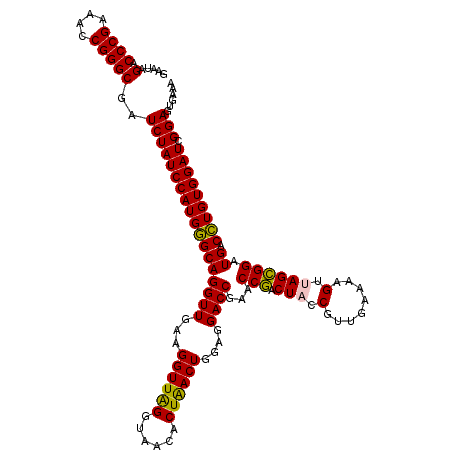
>pao.3201 6041964 120 - 6264404 GUAUAGACCCGAAACCGGGCGAUCUAUCCAUGAGCAGGUUGAAGGUUAGGUAACACUGACUGGAGGACCGAACCCACUCCCGUUGAAAAGGUAGGGGAUGACUUGUGGAUCGGAGUGAAA .....(.((((....)))))..((((((((..((((((((...((((((......))))))....))))...(((.((.((........)).))))).)).))..))))).)))...... ( -39.90) >pau.3340 6314883 120 - 6537648 GUAUAGACCCGAAACCGGGCGAUCUAUCCAUGAGCAGGUUGAAGGUUAGGUAACACUGACUGGAGGACCGAACCCACUCCCGUUGAAAAGGUAGGGGAUGACUUGUGGAUCGGAGUGAAA .....(.((((....)))))..((((((((..((((((((...((((((......))))))....))))...(((.((.((........)).))))).)).))..))))).)))...... ( -39.90) >pss.1919 3849832 120 - 6093698 GAAUAGACCCGAAACCGGGCGAUCUAUCCAUGGGCAGGUUGAAGGUUGGGUAACACUAACUGGAGGACCGAACCGACUACCGUUGAAAAGUUAGCGGAUGACCUGUGGAUCGGAGUGAAA .....(.((((....)))))..((((((((((((((((((...((((((......))))))....))))..........(((((((....))))))).)).))))))))).)))...... ( -40.20) >pel.69 5770496 120 + 5888780 GUAUAGACCCGAAACCGGGCGAUCUAUCCAUGGGCAGGUUGAAGGUUAGGUAACACUGACUGGAGGACCGAACCGACUACCGUUGAAAAGUUAGCGGAUGACCUGUGGAUCGGAGUGAAA .....(.((((....)))))..((((((((((((((((((...((((((......))))))....))))..........(((((((....))))))).)).))))))))).)))...... ( -40.60) >psp.1833 3633836 120 - 5928787 GAAUAGACCCGAAACCGGGCGAUCUAUCCAUGGGCAGGUUGAAGGUUGGGUAACACUAACUGGAGGACCGAACCGACUACCGUUGAAAAGUUAGCGGAUGACCUGUGGAUCGGAGUGAAA .....(.((((....)))))..((((((((((((((((((...((((((......))))))....))))..........(((((((....))))))).)).))))))))).)))...... ( -40.20) >pst.2031 3871833 120 - 6397126 GAAUAGACCCGAAACCGGGCGAUCUAUCCAUGGGCAGGUUGAAGGUUGGGUAACACUAACUGGAGGACCGAACCGACUACCGUUGAAAAGUUAGCGGAUGACCUGUGGAUCGGAGUGAAA .....(.((((....)))))..((((((((((((((((((...((((((......))))))....))))..........(((((((....))))))).)).))))))))).)))...... ( -40.20) >consensus GAAUAGACCCGAAACCGGGCGAUCUAUCCAUGGGCAGGUUGAAGGUUAGGUAACACUAACUGGAGGACCGAACCGACUACCGUUGAAAAGUUAGCGGAUGACCUGUGGAUCGGAGUGAAA .....(.((((....)))))..((((((((((((((((((...((((((......))))))....))))...(((.(((.(........).)))))).)).))))))))).)))...... (-39.02 = -37.97 + -1.05)
| Location | 6,042,004 – 6,042,123 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.37 |
| Mean single sequence MFE | -39.43 |
| Consensus MFE | -37.60 |
| Energy contribution | -37.55 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916670 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
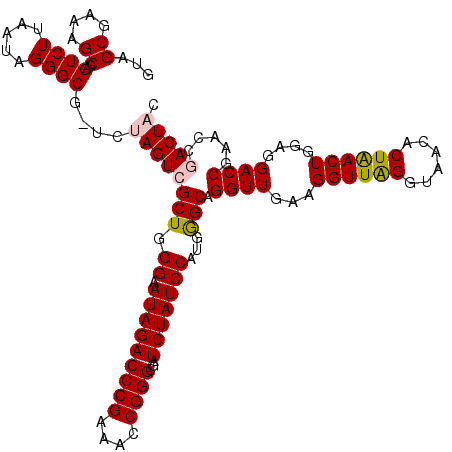
>pao.3201 6042004 119 - 6264404 GUAGCGAAAGCGAGUCUUAAUAGGGCG-UUUAGUCGCUGGGUAUAGACCCGAAACCGGGCGAUCUAUCCAUGAGCAGGUUGAAGGUUAGGUAACACUGACUGGAGGACCGAACCCACUCC ...((....))((((((....))((.(-((..((((((.((.(((((((((....))))...)))))))...)))........((((((......))))))....)))..))))))))). ( -38.30) >pau.3340 6314923 119 - 6537648 GUAGCGAAAGCGAGUCUUAAUAGGGCG-UUUAGUCGCUGGGUAUAGACCCGAAACCGGGCGAUCUAUCCAUGAGCAGGUUGAAGGUUAGGUAACACUGACUGGAGGACCGAACCCACUCC ...((....))((((((....))((.(-((..((((((.((.(((((((((....))))...)))))))...)))........((((((......))))))....)))..))))))))). ( -38.30) >pss.1919 3849872 119 - 6093698 GUAGCGAAAGCGAGUCUUAAUAGGGCG-UCUAGUCGCUGGGAAUAGACCCGAAACCGGGCGAUCUAUCCAUGGGCAGGUUGAAGGUUGGGUAACACUAACUGGAGGACCGAACCGACUAC ...((....))((((((.....)))).-))((((((((.(((.((((((((....))))...)))))))...))).((((...((((((......))))))....)))).....))))). ( -39.90) >pel.69 5770536 120 + 5888780 GUAGCGAAAGCGAGUCUUAAUAGGGCGCUUUAGUCGCUGGGUAUAGACCCGAAACCGGGCGAUCUAUCCAUGGGCAGGUUGAAGGUUAGGUAACACUGACUGGAGGACCGAACCGACUAC .(((((((((((..(((....))).)))))...))))))((.(((((((((....))))...))))))).(((...((((...((((((......))))))....))))...)))..... ( -40.30) >psp.1833 3633876 119 - 5928787 GUAGCGAAAGCGAGUCUUAAUAGGGCG-UCUAGUCGCUGGGAAUAGACCCGAAACCGGGCGAUCUAUCCAUGGGCAGGUUGAAGGUUGGGUAACACUAACUGGAGGACCGAACCGACUAC ...((....))((((((.....)))).-))((((((((.(((.((((((((....))))...)))))))...))).((((...((((((......))))))....)))).....))))). ( -39.90) >pst.2031 3871873 119 - 6397126 GUAGCGAAAGCGAGUCUUAAUAGGGCG-UCUAGUCGCUGGGAAUAGACCCGAAACCGGGCGAUCUAUCCAUGGGCAGGUUGAAGGUUGGGUAACACUAACUGGAGGACCGAACCGACUAC ...((....))((((((.....)))).-))((((((((.(((.((((((((....))))...)))))))...))).((((...((((((......))))))....)))).....))))). ( -39.90) >consensus GUAGCGAAAGCGAGUCUUAAUAGGGCG_UCUAGUCGCUGGGAAUAGACCCGAAACCGGGCGAUCUAUCCAUGGGCAGGUUGAAGGUUAGGUAACACUAACUGGAGGACCGAACCGACUAC ...((....))..((((.....))))....((((((((.((.(((((((((....))))...)))))))...))).((((...((((((......))))))....)))).....))))). (-37.60 = -37.55 + -0.05)

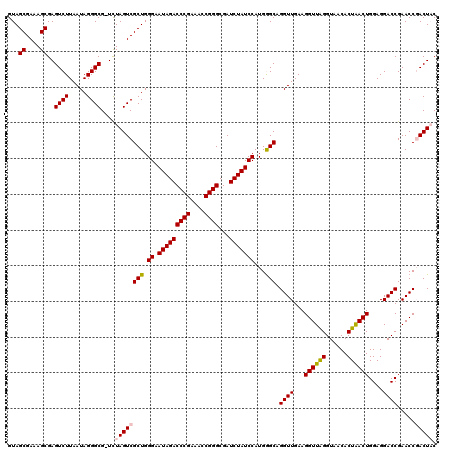
| Location | 6,042,044 – 6,042,163 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.37 |
| Mean single sequence MFE | -42.78 |
| Consensus MFE | -40.38 |
| Energy contribution | -40.22 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.94 |
| SVM decision value | 3.53 |
| SVM RNA-class probability | 0.999341 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.3201 6042044 119 - 6264404 GACUUAUAUUCAGUGGCAAGCUUAACCGUAUAGGGUAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCG-UUUAGUCGCUGGGUAUAGACCCGAAACCGGGCGAUCUAUCCAUGAGCAGGUU (.(((((...(((((((..(((((.((.....)).)))))...((....))..((((.....)))).-....)))))))((.(((((((((....))))...)))))))))))))..... ( -40.80) >pau.3340 6314963 119 - 6537648 GACUUAUAUUCAGUGGCAAGCUUAACCGUAUAGGGUAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCG-UUUAGUCGCUGGGUAUAGACCCGAAACCGGGCGAUCUAUCCAUGAGCAGGUU (.(((((...(((((((..(((((.((.....)).)))))...((....))..((((.....)))).-....)))))))((.(((((((((....))))...)))))))))))))..... ( -40.80) >pel.69 5770576 120 + 5888780 GACUUAUAUUCAGUGGCGAGCUUAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCGCUUUAGUCGCUGGGUAUAGACCCGAAACCGGGCGAUCUAUCCAUGGGCAGGUU ....(((((((((((((..((((..((.....))..))))((.((....))..((((.....))))))....)))))))))))))(.((((....)))))(((((..(....)..))))) ( -44.00) >pf5.3378 6385201 119 - 7074893 GACUUAUUUUCAGUGGCAAGCUUAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCG-UCUAGUCGCUGGGAAUAGACCCGAAACCGGGCGAUCUAUCCAUGGGCAGGUU ....((((..(((((((..((((..((.....))..))))...((....))((((((.....)))).-))..)))))))..))))(.((((....)))))(((((..(....)..))))) ( -43.70) >pfo.3002 5728821 119 - 6438405 GACUUAUUUUCAGUGGCGAGCUUAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCG-UCUAGUCGCUGGGAAUAGACCCGAAACCGGGCGAUCUAUCCAUGGGCAGGUU ....((((..(((((((..((((..((.....))..))))...((....))((((((.....)))).-))..)))))))..))))(.((((....)))))(((((..(....)..))))) ( -43.70) >pst.2031 3871913 119 - 6397126 GACUUAUUUUCAGUGGCGAGCUUAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCG-UCUAGUCGCUGGGAAUAGACCCGAAACCGGGCGAUCUAUCCAUGGGCAGGUU ....((((..(((((((..((((..((.....))..))))...((....))((((((.....)))).-))..)))))))..))))(.((((....)))))(((((..(....)..))))) ( -43.70) >consensus GACUUAUAUUCAGUGGCAAGCUUAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCG_UCUAGUCGCUGGGAAUAGACCCGAAACCGGGCGAUCUAUCCAUGGGCAGGUU ....(((((((((((((..((((..((.....))..))))...((....))..((((.....))))......)))))))))))))(.((((....)))))(((((.((....)).))))) (-40.38 = -40.22 + -0.17)

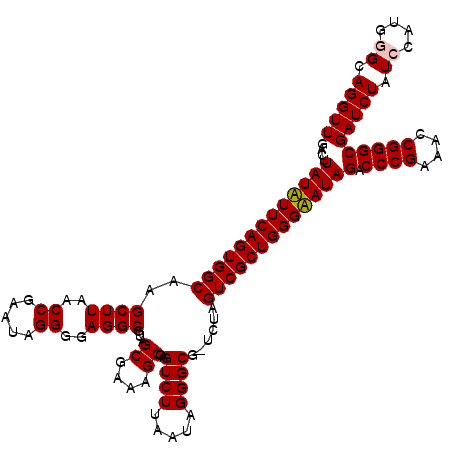
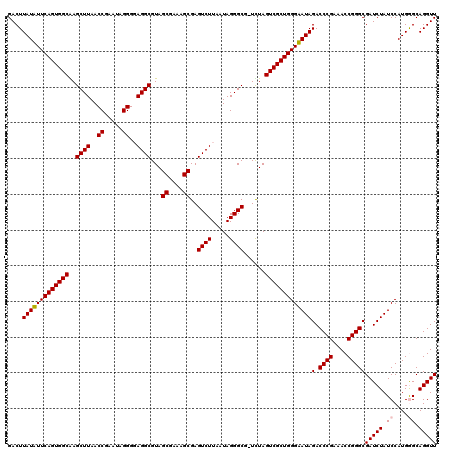
Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:04:19 2007