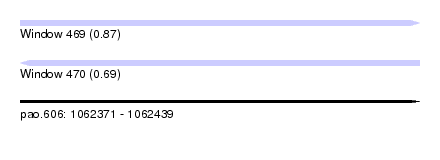
| Sequence ID | pao.606 |
|---|---|
| Location | 1,062,371 – 1,062,439 |
| Length | 68 |
| Max. P | 0.869633 |

| Location | 1,062,371 – 1,062,439 |
|---|---|
| Length | 68 |
| Sequences | 6 |
| Columns | 68 |
| Reading direction | forward |
| Mean pairwise identity | 82.49 |
| Mean single sequence MFE | -20.38 |
| Consensus MFE | -14.72 |
| Energy contribution | -14.92 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.869633 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.606 1062371 68 + 6264404 GACACCUCUUUCACGGCGAGGAUUCUCGCCUAAAUCGGUGUCCGGGAUCAGUAGACCACUACACUGCC ((((((..(((...((((((....)))))).)))..))))))..((....((((....))))....)) ( -22.00) >pau.2426 1949810 68 - 6537648 GACACCUCUUUCACGGCGAGGAUUCUCGCCUAAAUCGGUGUCCGGGAUCAGUAGACCACUACACUGCC ((((((..(((...((((((....)))))).)))..))))))..((....((((....))))....)) ( -22.00) >pss.2296 4534894 62 + 6093698 GACACCUCUAUC-UGGCGA-UAUUCUCGCCUAAAUGAGUGUCCGGAGUCAUUAGACCACUACAC---- (((((.((....-.(((((-.....))))).....))))))).(..(((....)))..).....---- ( -18.20) >ppk.2543 1167678 62 - 6181863 GACACCUCUUUG-GGGCGAGCAUUCUCGCCUAAAUGAGUGUCCGGUUUCAUUAGACCACUACA----- (((((.((....-(((((((....)))))))....))))))).((((......))))......----- ( -23.80) >pfo.1747 3013580 61 - 6438405 GACACCUCUAUC-UGGCGAGCAUUUUCGCCUAAAUGGGUGUCCGGUUUCAUUAGACCACUGC------ (((((((.....-.((((((....)))))).....))))))).((((......)))).....------ ( -22.70) >pst.2801 1086691 62 - 6397126 GACACCUCUAUC-UGGCGA-CAUUCUUGCCUAAGUGAGUGUCCGGAAUCAUUAGACCAGUACAA---- (((((.((....-.(((((-.....))))).....))))))).((..((....)))).......---- ( -13.60) >consensus GACACCUCUAUC_UGGCGAGCAUUCUCGCCUAAAUGAGUGUCCGGAAUCAUUAGACCACUACAC____ (((((.((......((((((....)))))).....))))))).((..........))........... (-14.72 = -14.92 + 0.20)

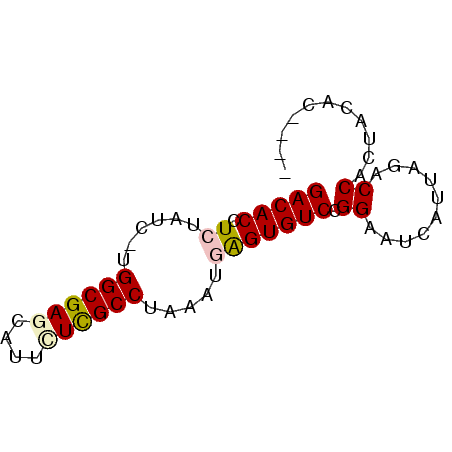
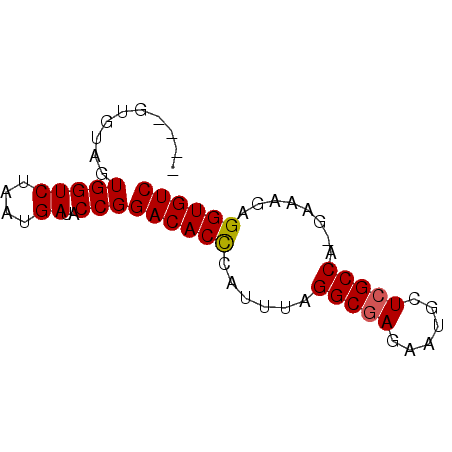
| Location | 1,062,371 – 1,062,439 |
|---|---|
| Length | 68 |
| Sequences | 6 |
| Columns | 68 |
| Reading direction | reverse |
| Mean pairwise identity | 82.49 |
| Mean single sequence MFE | -21.05 |
| Consensus MFE | -14.87 |
| Energy contribution | -14.79 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.693592 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
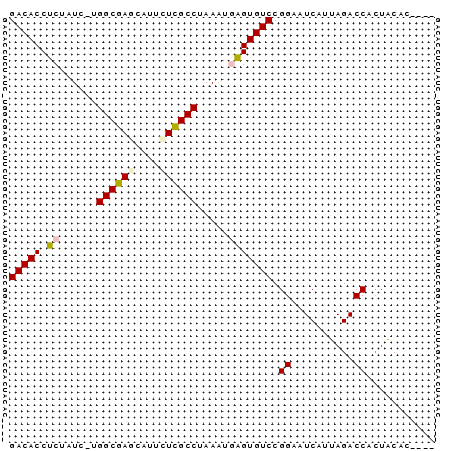
>pao.606 1062371 68 - 6264404 GGCAGUGUAGUGGUCUACUGAUCCCGGACACCGAUUUAGGCGAGAAUCCUCGCCGUGAAAGAGGUGUC ((....((((....)))).....)).((((((..(((.((((((....))))))...)))..)))))) ( -26.80) >pau.2426 1949810 68 + 6537648 GGCAGUGUAGUGGUCUACUGAUCCCGGACACCGAUUUAGGCGAGAAUCCUCGCCGUGAAAGAGGUGUC ((....((((....)))).....)).((((((..(((.((((((....))))))...)))..)))))) ( -26.80) >pss.2296 4534894 62 - 6093698 ----GUGUAGUGGUCUAAUGACUCCGGACACUCAUUUAGGCGAGAAUA-UCGCCA-GAUAGAGGUGUC ----.....(..(((....)))..).(((((((((((.(((((.....-))))))-))).)).))))) ( -20.70) >ppk.2543 1167678 62 + 6181863 -----UGUAGUGGUCUAAUGAAACCGGACACUCAUUUAGGCGAGAAUGCUCGCCC-CAAAGAGGUGUC -----.....((((........))))((((((..(((.((((((....)))))).-.)))..)))))) ( -20.50) >pfo.1747 3013580 61 + 6438405 ------GCAGUGGUCUAAUGAAACCGGACACCCAUUUAGGCGAAAAUGCUCGCCA-GAUAGAGGUGUC ------....((((........))))(((((((((((.(((((......))))))-))).).)))))) ( -19.90) >pst.2801 1086691 62 + 6397126 ----UUGUACUGGUCUAAUGAUUCCGGACACUCACUUAGGCAAGAAUG-UCGCCA-GAUAGAGGUGUC ----.(((.((((((....))..)))))))........((((....((-((....-))))....)))) ( -11.60) >consensus ____GUGUAGUGGUCUAAUGAUACCGGACACCCAUUUAGGCGAGAAUGCUCGCCA_GAAAGAGGUGUC ..........(((((....))..)))((((((......(((((......)))))........)))))) (-14.87 = -14.79 + -0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:11:50 2007