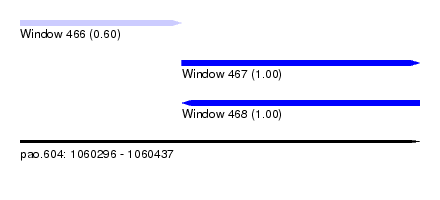
| Sequence ID | pao.604 |
|---|---|
| Location | 1,060,296 – 1,060,437 |
| Length | 141 |
| Max. P | 0.999993 |

| Location | 1,060,296 – 1,060,353 |
|---|---|
| Length | 57 |
| Sequences | 6 |
| Columns | 58 |
| Reading direction | forward |
| Mean pairwise identity | 72.52 |
| Mean single sequence MFE | -11.87 |
| Consensus MFE | -3.39 |
| Energy contribution | -3.75 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.29 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.597614 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.604 1060296 57 + 6264404 AAAAAA-UUCACGAGGGGUGUUGUUUUUACGUUAGAAAUCAGUAUUAUACGCCUCGCG ......-....(((((.((((......(((...........)))..)))).))))).. ( -12.20) >pel.2353 1507321 56 - 5888780 --AAAAUUUCUGAUGGGGUGUUGAUUUCCUCUGAGAAAUCAGUAAUAUACGCGCCAUC --.........(((((.((((..(((....((((....)))).)))..)))).))))) ( -17.50) >pss.851 1604259 52 + 6093698 ------UUUCAAAAAAGGUGUUGAAUUACUCAUAAAAAUCAGUAAUAUACGCGCCACA ------..........(((((((.((((((.((....)).)))))).)).)))))... ( -9.60) >pf5.2892 1581248 52 - 7074893 ------UUUCAUAUAGGGUGUUGAAUUCCCGUUAGAAAUCAGUAUUAUACGCGCCACC ------..........((((((((.(((......))).)))(((...))))))))... ( -8.10) >pfo.2611 1478732 52 - 6438405 ------UUUCAAAUUAGGUGUUGAAUAGUCCUUAAAAAUCAGUAUUAUACGCGCCACC ------..........(((((.(.(((((.((........)).))))).))))))... ( -8.00) >psp.2196 1614043 52 - 5928787 ------UUUUGAAAAAGGUGUUGAAUUACUGAUAAAAAUCAGUAAUAUACGCGCCACA ------..........(((((((.(((((((((....))))))))).)).)))))... ( -15.80) >consensus ______UUUCAAAAAAGGUGUUGAAUUACCCUUAAAAAUCAGUAAUAUACGCGCCACC ................((((((((.((........)).)))(((...))))))))... ( -3.39 = -3.75 + 0.36)

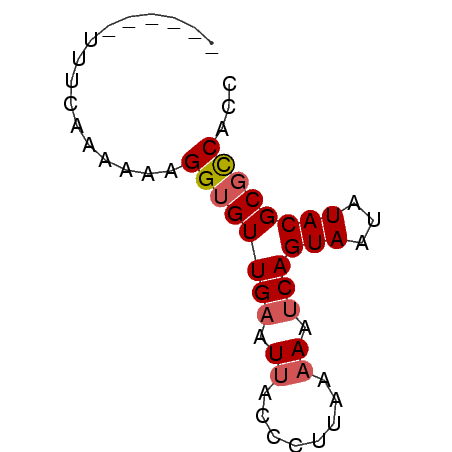

| Location | 1,060,353 – 1,060,437 |
|---|---|
| Length | 84 |
| Sequences | 6 |
| Columns | 85 |
| Reading direction | forward |
| Mean pairwise identity | 93.75 |
| Mean single sequence MFE | -31.53 |
| Consensus MFE | -31.00 |
| Energy contribution | -31.67 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.75 |
| Structure conservation index | 0.98 |
| SVM decision value | 5.76 |
| SVM RNA-class probability | 0.999993 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.604 1060353 84 + 6264404 UU-GGGUCGUUAGCUCAGUCGGUAGAGCAGUUGGCUUUUAACCAAUUGGUCGUAGGUUCGAAUCCUACACGACCCACCAUUUUCU .(-(((((((..............((.(((((((.......))))))).))(((((.......)))))))))))))......... ( -30.80) >pau.2428 1946468 84 - 6537648 UU-GGGUCGUUAGCUCAGUCGGUAGAGCAGUUGGCUUUUAACCAAUUGGUCGUAGGUUCGAAUCCUACACGACCCACCAUUUUCG .(-(((((((..............((.(((((((.......))))))).))(((((.......)))))))))))))......... ( -30.80) >pel.2694 912544 80 - 5888780 UCCGGGUCGUUAGCUCAGUUGGUAGAGCAGUUGGCUUUUAACCAAUUGGUCGUAGGUUCGAAUCCUACACGACCCACCAU----- ...(((((((..(..(((((((((((((.....)))))..))))))))..)(((((.......)))))))))))).....----- ( -34.00) >ppk.352 733545 80 + 6181863 UCCGGGUCGUUAGCUCAGUUGGUAGAGCAGUUGGCUUUUAACCAAUUGGUCGUAGGUUCGAAUCCUACACGACCCACCAU----- ...(((((((..(..(((((((((((((.....)))))..))))))))..)(((((.......)))))))))))).....----- ( -34.00) >pst.498 898969 84 + 6397126 UU-GGGUCGUUAGCUCAGUUGGUAGAGCAGUUGGCUUUUAACCAAUUGGUCGUAGGUUCGAAUCCCACACGACCCACCAUUUUUG .(-(((((((..(..(((((((((((((.....)))))..))))))))..)((.((.......)).))))))))))......... ( -29.80) >psp.432 871060 84 + 5928787 UU-GGGUCGUUAGCUCAGUUGGUAGAGCAGUUGGCUUUUAACCAAUUGGUCGUAGGUUCGAAUCCCACACGACCCACCAUUUUUG .(-(((((((..(..(((((((((((((.....)))))..))))))))..)((.((.......)).))))))))))......... ( -29.80) >consensus UU_GGGUCGUUAGCUCAGUUGGUAGAGCAGUUGGCUUUUAACCAAUUGGUCGUAGGUUCGAAUCCUACACGACCCACCAUUUU_G ...(((((((..(..(((((((((((((.....)))))..))))))))..)(((((.......)))))))))))).......... (-31.00 = -31.67 + 0.67)

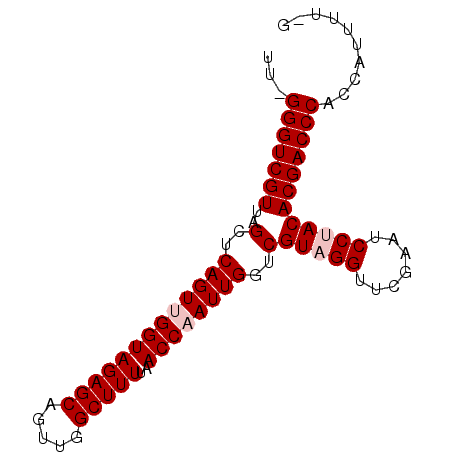
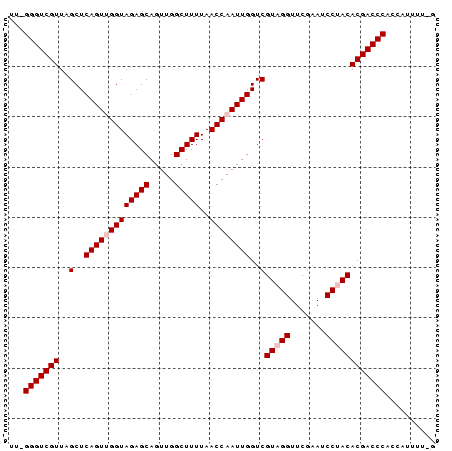
| Location | 1,060,353 – 1,060,437 |
|---|---|
| Length | 84 |
| Sequences | 6 |
| Columns | 85 |
| Reading direction | reverse |
| Mean pairwise identity | 93.75 |
| Mean single sequence MFE | -28.97 |
| Consensus MFE | -27.39 |
| Energy contribution | -27.17 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.51 |
| Structure conservation index | 0.95 |
| SVM decision value | 5.03 |
| SVM RNA-class probability | 0.999969 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.604 1060353 84 - 6264404 AGAAAAUGGUGGGUCGUGUAGGAUUCGAACCUACGACCAAUUGGUUAAAAGCCAACUGCUCUACCGACUGAGCUAACGACCC-AA .........(((((((((((((.......)))))......(((((.....)))))..((((........))))..)))))))-). ( -29.30) >pau.2428 1946468 84 + 6537648 CGAAAAUGGUGGGUCGUGUAGGAUUCGAACCUACGACCAAUUGGUUAAAAGCCAACUGCUCUACCGACUGAGCUAACGACCC-AA .........(((((((((((((.......)))))......(((((.....)))))..((((........))))..)))))))-). ( -29.30) >pel.2694 912544 80 + 5888780 -----AUGGUGGGUCGUGUAGGAUUCGAACCUACGACCAAUUGGUUAAAAGCCAACUGCUCUACCAACUGAGCUAACGACCCGGA -----....(((((((((((((.......)))))......(((((.....)))))..((((........))))..)))))))).. ( -28.60) >ppk.352 733545 80 - 6181863 -----AUGGUGGGUCGUGUAGGAUUCGAACCUACGACCAAUUGGUUAAAAGCCAACUGCUCUACCAACUGAGCUAACGACCCGGA -----....(((((((((((((.......)))))......(((((.....)))))..((((........))))..)))))))).. ( -28.60) >pst.498 898969 84 - 6397126 CAAAAAUGGUGGGUCGUGUGGGAUUCGAACCUACGACCAAUUGGUUAAAAGCCAACUGCUCUACCAACUGAGCUAACGACCC-AA .........(((((((((((((.......)))))......(((((.....)))))..((((........))))..)))))))-). ( -29.00) >psp.432 871060 84 - 5928787 CAAAAAUGGUGGGUCGUGUGGGAUUCGAACCUACGACCAAUUGGUUAAAAGCCAACUGCUCUACCAACUGAGCUAACGACCC-AA .........(((((((((((((.......)))))......(((((.....)))))..((((........))))..)))))))-). ( -29.00) >consensus C_AAAAUGGUGGGUCGUGUAGGAUUCGAACCUACGACCAAUUGGUUAAAAGCCAACUGCUCUACCAACUGAGCUAACGACCC_AA ..........((((((((((((.......)))))......(((((.....)))))..((((........))))..)))))))... (-27.39 = -27.17 + -0.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:11:48 2007