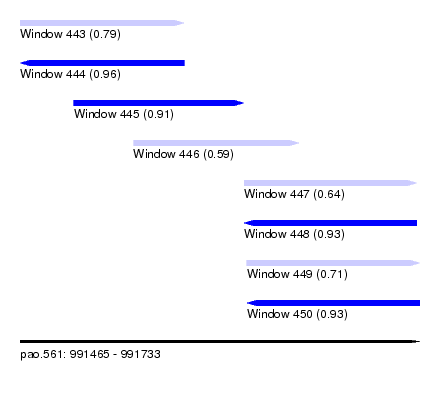
| Sequence ID | pao.561 |
|---|---|
| Location | 991,465 – 991,733 |
| Length | 268 |
| Max. P | 0.955344 |

| Location | 991,465 – 991,575 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 78.64 |
| Mean single sequence MFE | -36.80 |
| Consensus MFE | -32.64 |
| Energy contribution | -30.37 |
| Covariance contribution | -2.27 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.790343 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.561 991465 110 + 6264404 -ACGCACUCAUAGCUCAGCUGGAUAGAGUACUCGGCUACGAACCGAGCGGUCGGAGGUUCGAAUCCUCCUGAGUGCGCCAUAUUUCGA-AGGGUUCCUCU-----CGUCAGGGGCUC -..(((((((((((...))))....((.(.(((((.......))))).).))(((((.......))))))))))))(((......(((-..(....)..)-----)).....))).. ( -39.90) >pfo.2515 1617487 101 - 6438405 AAUGCACUCGUAGCUCAGCUGGAUAGAGUACUCGGCUACGAACCGAGCGGUCACAGGUUCGAAUCCUGUCGAGUGCACCA---UUUAAGAG--------U-----CAAUUGCAGCGA ..((((....(..(((...(((((.(.(((((((((..((((((...........))))))......))))))))).).)---)))).)))--------.-----.)..)))).... ( -29.20) >pf5.2742 1861247 101 - 7074893 AAUGCACUCGUAGCUCAGCUGGAUAGAGUACUCGGCUACGAACCGAGCGGUCACAGGUUCGAAUCCUGUCGAGUGCACCA---UUUAAGAG--------U-----UAGUUACAGUUA .....(((.((((((.((((((((.(.(((((((((..((((((...........))))))......))))))))).).)---)))...))--------)-----)))))))))).. ( -30.90) >pel.2216 1732103 107 - 5888780 CACGGACUCAUAGCUCAGCUGGAUAGAGUACUCGGCUACGAACCGAGCGGUCGGAGGUUCGAAUCCUCCUGAGUCCGCCAC-UUUUGA-AG--------UGGCUUUGCUUACAAGGC ...(((((((((((...))))....((.(.(((((.......))))).).))(((((.......))))))))))))(((((-(.....-))--------))))...(((.....))) ( -40.10) >pau.2472 1877523 110 - 6537648 -ACGCACUCAUAGCUCAGCUGGAUAGAGUACUCGGCUACGAACCGAGCGGUCGGAGGUUCGAAUCCUCCUGAGUGCGCCAUAUUUCGA-AGGGUUCCUCU-----CGUCAGGGGCUC -..(((((((((((...))))....((.(.(((((.......))))).).))(((((.......))))))))))))(((......(((-..(....)..)-----)).....))).. ( -39.90) >ppk.2577 1102185 107 - 6181863 CACGGACUCAUAGCUCAGCUGGAUAGAGUACUCGGCUACGAACCGAGCGGUCGCAGGUUCGAAUCCUGCUGAGUCCGCCAC-UUUUGA-AG--------UGGCUUUGUUUGCAAGGC ..(((((.....(((((((.((((..(((.....))).((((((..((....)).)))))).)))).)))))))..(((((-(.....-))--------))))...)))))...... ( -40.80) >consensus _ACGCACUCAUAGCUCAGCUGGAUAGAGUACUCGGCUACGAACCGAGCGGUCGCAGGUUCGAAUCCUCCUGAGUGCGCCA__UUUUGA_AG________U_____CGUUUGCAGCGC ..((((((((((((...))))....((.(.(((((.......))))).).))(((((.......)))))))))))))........................................ (-32.64 = -30.37 + -2.27)

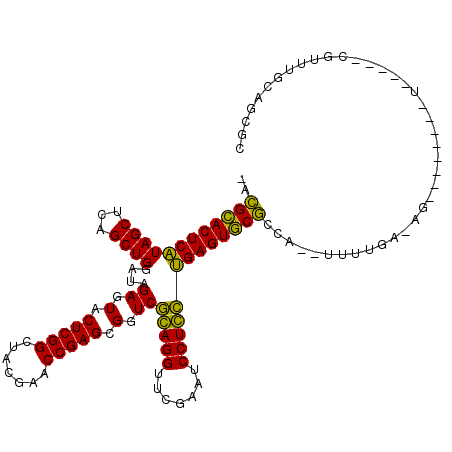
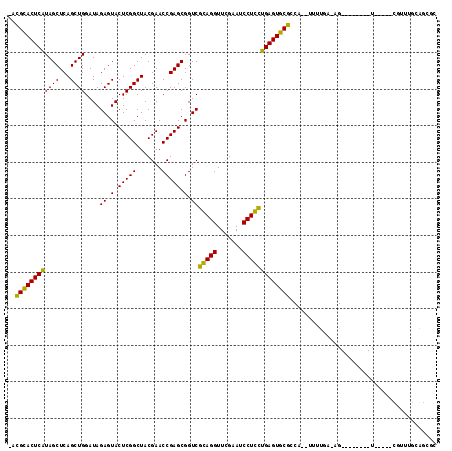
| Location | 991,465 – 991,575 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 78.64 |
| Mean single sequence MFE | -37.90 |
| Consensus MFE | -36.44 |
| Energy contribution | -34.17 |
| Covariance contribution | -2.27 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.955344 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.561 991465 110 - 6264404 GAGCCCCUGACG-----AGAGGAACCCU-UCGAAAUAUGGCGCACUCAGGAGGAUUCGAACCUCCGACCGCUCGGUUCGUAGCCGAGUACUCUAUCCAGCUGAGCUAUGAGUGCGU- ..(((.......-----.((((....))-)).......)))((((((((((((.......)))))((..(((((((.....)))))))..))...............)))))))..- ( -39.36) >pfo.2515 1617487 101 + 6438405 UCGCUGCAAUUG-----A--------CUCUUAAA---UGGUGCACUCGACAGGAUUCGAACCUGUGACCGCUCGGUUCGUAGCCGAGUACUCUAUCCAGCUGAGCUACGAGUGCAUU ............-----.--------........---.(((((((((((((((.......)))))((..(((((((.....)))))))..))...............)))))))))) ( -32.10) >pf5.2742 1861247 101 + 7074893 UAACUGUAACUA-----A--------CUCUUAAA---UGGUGCACUCGACAGGAUUCGAACCUGUGACCGCUCGGUUCGUAGCCGAGUACUCUAUCCAGCUGAGCUACGAGUGCAUU ............-----.--------........---.(((((((((((((((.......)))))((..(((((((.....)))))))..))...............)))))))))) ( -32.10) >pel.2216 1732103 107 + 5888780 GCCUUGUAAGCAAAGCCA--------CU-UCAAAA-GUGGCGGACUCAGGAGGAUUCGAACCUCCGACCGCUCGGUUCGUAGCCGAGUACUCUAUCCAGCUGAGCUAUGAGUCCGUG .........((...((((--------((-.....)-)))))((((((((((((.......)))))((..(((((((.....)))))))..))...............))))))))). ( -42.40) >pau.2472 1877523 110 + 6537648 GAGCCCCUGACG-----AGAGGAACCCU-UCGAAAUAUGGCGCACUCAGGAGGAUUCGAACCUCCGACCGCUCGGUUCGUAGCCGAGUACUCUAUCCAGCUGAGCUAUGAGUGCGU- ..(((.......-----.((((....))-)).......)))((((((((((((.......)))))((..(((((((.....)))))))..))...............)))))))..- ( -39.36) >ppk.2577 1102185 107 + 6181863 GCCUUGCAAACAAAGCCA--------CU-UCAAAA-GUGGCGGACUCAGCAGGAUUCGAACCUGCGACCGCUCGGUUCGUAGCCGAGUACUCUAUCCAGCUGAGCUAUGAGUCCGUG .....((.......((((--------((-.....)-)))))((((((((((((.......)))))((..(((((((.....)))))))..))...............))))))))). ( -42.10) >consensus GAGCUGCAAACA_____A________CU_UCAAAA__UGGCGCACUCAGCAGGAUUCGAACCUCCGACCGCUCGGUUCGUAGCCGAGUACUCUAUCCAGCUGAGCUAUGAGUGCGU_ .......................................((((((((((((((.......)))))((..(((((((.....)))))))..))...............))))))))). (-36.44 = -34.17 + -2.27)
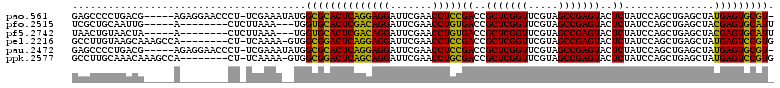
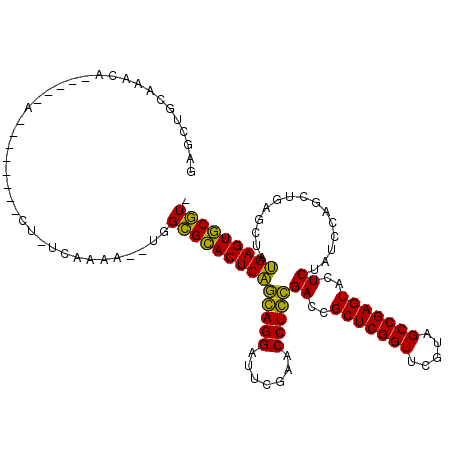
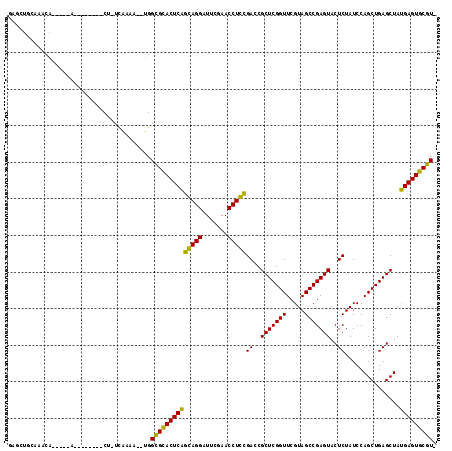
| Location | 991,501 – 991,615 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 68.42 |
| Mean single sequence MFE | -26.85 |
| Consensus MFE | -16.72 |
| Energy contribution | -15.12 |
| Covariance contribution | -1.60 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.910307 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
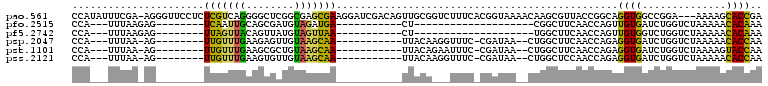
>pao.561 991501 114 + 6264404 ACGAACCGAGCGGUCGGAGGUUCGAAUCCUCCUGAGUGCGCCAUAUUUCGA-AGGGUUCCUCUCGUCAGGGGCUCGGCGAGCGAAGGAUCGACAGUUGCGGUCUUUCACGGUAAA ....((((.(((.(((((((.......))))).)).))).......((((.-.((((((((......))))))))..)))).(((((((((.(....)))))))))).))))... ( -45.60) >pfo.2515 1617524 75 - 6438405 ACGAACCGAGCGGUCACAGGUUCGAAUCCUGUCGAGUGCACCA---UUUAAGAG--------UCAAUUGCAGCGAUGUAGAUGA-----------CU------------------ .........(((.(((((((.......))))).)).)))....---......((--------(((.(((((....))))).)))-----------))------------------ ( -21.80) >pf5.2742 1861284 75 - 7074893 ACGAACCGAGCGGUCACAGGUUCGAAUCCUGUCGAGUGCACCA---UUUAAGAG--------UUAGUUACAGUUAUGUAGUUAA-----------CU------------------ .........(((.(((((((.......))))).)).)))....---......((--------(((..((((....))))..)))-----------))------------------ ( -20.70) >psp.2047 1882464 90 - 5928787 ACGAACCGAGCGGUCACAGGUUCGAAUCCUGUCGAGUGCACCA---UUUAA-AG--------UUGUUUGAAGAGUUGUAAGCAA-----------UUACAAGGUUUC-CGAUAA- .........(((.(((((((.......))))).)).)))....---.....-..--------(((((.((((..((((((....-----------))))))..))))-.)))))- ( -23.80) >pst.1101 2015043 90 + 6397126 ACGAACCGAGCGGUCACAGGUUCGAAUCCUGUCGAGUGCACCA---UUUAA-AG--------UUGUUUGAAGCGCUGUAAGCAA-----------UUACAGAAUUUC-CGAUAA- .........(((.(((((((.......))))).)).)))....---.....-..--------(((((.((((..((((((....-----------))))))..))))-.)))))- ( -25.30) >pss.2121 1860575 90 - 6093698 ACGAACCGAGCGGUCACAGGUUCGAAUCCUGUCGAGUGCACCA---UUUAA-AG--------UUGUUUGAAGUGUUGUAAGCAA-----------UUACAAGGUUUC-CGAUAA- .........(((.(((((((.......))))).)).)))....---.....-..--------(((((.((((..((((((....-----------))))))..))))-.)))))- ( -23.90) >consensus ACGAACCGAGCGGUCACAGGUUCGAAUCCUGUCGAGUGCACCA___UUUAA_AG________UUGUUUGAAGCGAUGUAAGCAA___________UUACA___UUUC_CGAUAA_ .........(((.(((((((.......))))).)).))).......................(((((((........)))))))............................... (-16.72 = -15.12 + -1.60)

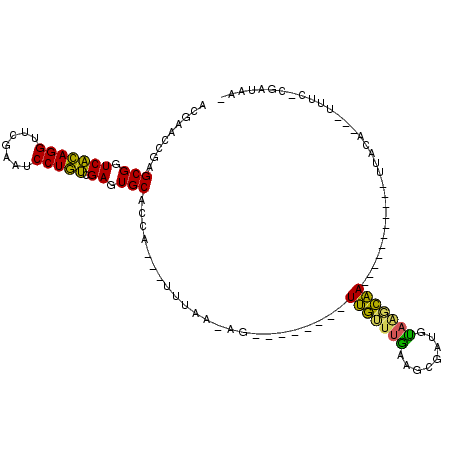
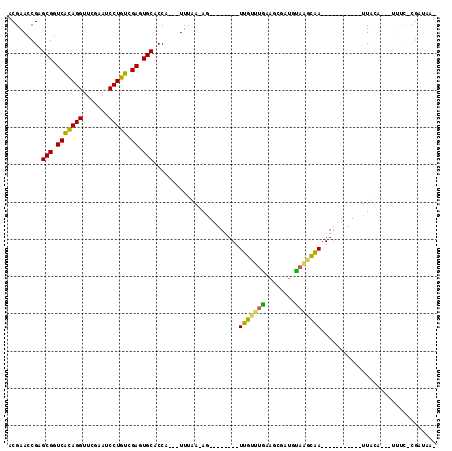
| Location | 991,541 – 991,652 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 60.47 |
| Mean single sequence MFE | -24.72 |
| Consensus MFE | -6.19 |
| Energy contribution | -4.56 |
| Covariance contribution | -1.63 |
| Combinations/Pair | 1.73 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.25 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.593108 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.561 991541 111 + 6264404 CCAUAUUUCGA-AGGGUUCCUCUCGUCAGGGGCUCGGCGAGCGAAGGAUCGACAGUUGCGGUCUUUCACGGUAAAACAAGCGUUACCGGCAGGUGGCCGGA---AAAAGCACCGA ......((((.-.((((((((......))))))))..)))).(((((((((.(....)))))))))).((((..(((....))).(((((.....))))).---......)))). ( -40.50) >pfo.2515 1617564 73 - 6438405 CCA---UUUAAGAG--------UCAAUUGCAGCGAUGUAGAUGA-----------CU--------------------CGGCUUCAACCAGUUGUGAUCUGGUCUAAAAACACAAA ...---.....(((--------(((.(((((....))))).)))-----------))--------------------).......(((((.......)))))............. ( -18.30) >pf5.2742 1861324 73 - 7074893 CCA---UUUAAGAG--------UUAGUUACAGUUAUGUAGUUAA-----------CU--------------------UGGCUUCAACCAGUUGUGGUCUGGUCUAAAAACACAAA (((---.....(((--------(((..((((....))))..)))-----------))--------------------)((((.(((....))).))))))).............. ( -16.20) >psp.2047 1882504 89 - 5928787 CCA---UUUAA-AG--------UUGUUUGAAGAGUUGUAAGCAA-----------UUACAAGGUUUC-CGAUAA--CUGGCUUCAACCAGAGGUGAUCUGGUCUAAAAACACCAA ...---.....-((--------(((((.((((..((((((....-----------))))))..))))-.)))))--)).......((((((.....))))))............. ( -23.30) >pst.1101 2015083 89 + 6397126 CCA---UUUAA-AG--------UUGUUUGAAGCGCUGUAAGCAA-----------UUACAGAAUUUC-CGAUAA--CUGGCUUCAACCAGAGGUGAUCUGGUCUAAAAGUACCAA ...---.....-((--------(((((.((((..((((((....-----------))))))..))))-.)))))--)).((((..((((((.....))))))....))))..... ( -26.60) >pss.2121 1860615 89 - 6093698 CCA---UUUAA-AG--------UUGUUUGAAGUGUUGUAAGCAA-----------UUACAAGGUUUC-CGAUAA--CUGGCUCCAACCAGAGGUGAUCUGGUCUAAAAACACCAA ...---.....-((--------(((((.((((..((((((....-----------))))))..))))-.)))))--)).......((((((.....))))))............. ( -23.40) >consensus CCA___UUUAA_AG________UUGUUUGAAGCGAUGUAAGCAA___________UUACA___UUUC_CGAUAA__CUGGCUUCAACCAGAGGUGAUCUGGUCUAAAAACACCAA ......................(((((((........)))))))...............................................((((..............)))).. ( -6.19 = -4.56 + -1.63)
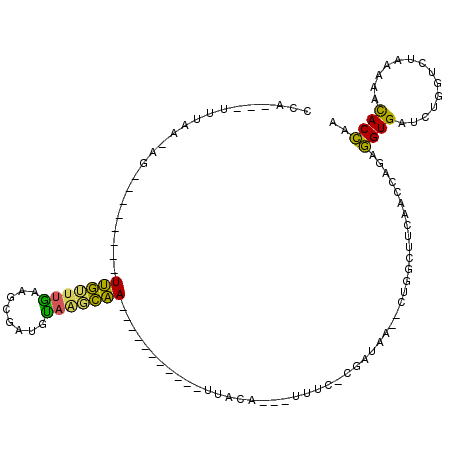
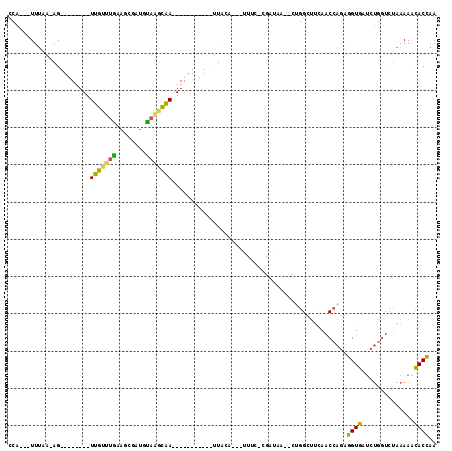
| Location | 991,615 – 991,731 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.37 |
| Mean single sequence MFE | -39.08 |
| Consensus MFE | -35.40 |
| Energy contribution | -32.49 |
| Covariance contribution | -2.91 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.641301 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
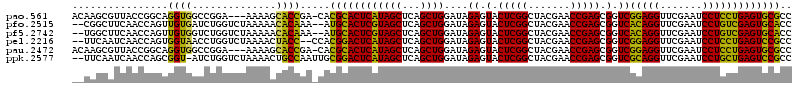
>pao.561 991615 116 + 6264404 ACAAGCGUUACCGGCAGGUGGCCGGA---AAAAGCACCGA-CACGCACUCAUAGCUCAGCUGGAUAGAGUACUCGGCUACGAACCGAGCGGUCGGAGGUUCGAAUCCUCCUGAGUGCGCC ....((....(((((.....))))).---....(((((((-(.(((((((.((((...))))....))))..((((.......))))))))))(((((.......))))).).)))))). ( -44.00) >pfo.2515 1617599 116 - 6438405 --CGGCUUCAACCAGUUGUGAUCUGGUCUAAAAACACAAA--AUGCACUCGUAGCUCAGCUGGAUAGAGUACUCGGCUACGAACCGAGCGGUCACAGGUUCGAAUCCUGUCGAGUGCACC --........(((((.......))))).............--.(((((((((((...(((((..((...))..))))).((((((...........))))))....))).)))))))).. ( -34.00) >pf5.2742 1861359 116 - 7074893 --UGGCUUCAACCAGUUGUGGUCUGGUCUAAAAACACAAA--AUGCACUCGUAGCUCAGCUGGAUAGAGUACUCGGCUACGAACCGAGCGGUCACAGGUUCGAAUCCUGUCGAGUGCACC --.((((...((((....))))..))))............--.(((((((((((...(((((..((...))..))))).((((((...........))))))....))).)))))))).. ( -35.30) >pel.2216 1732213 116 - 5888780 --UUCAAUCAACCAGUGGUAACCUGGUCUAAAACUACC--CCACGGACUCAUAGCUCAGCUGGAUAGAGUACUCGGCUACGAACCGAGCGGUCGGAGGUUCGAAUCCUCCUGAGUCCGCC --........(((((.(....))))))...........--...((((((((((((...))))....((.(.(((((.......))))).).))(((((.......))))))))))))).. ( -37.50) >pau.2472 1877673 116 - 6537648 ACAAGCGUUACCGGCAGGUGGCCGGA---AAAAGCACCGA-CACGCACUCAUAGCUCAGCUGGAUAGAGUACUCGGCUACGAACCGAGCGGUCGGAGGUUCGAAUCCUCCUGAGUGCGCC ....((....(((((.....))))).---....(((((((-(.(((((((.((((...))))....))))..((((.......))))))))))(((((.......))))).).)))))). ( -44.00) >ppk.2577 1102295 117 - 6181863 --UUCAAUCAACCAGCGGU-AUCUGGUCUAAAACUGCCAAUUGCGGACUCAUAGCUCAGCUGGAUAGAGUACUCGGCUACGAACCGAGCGGUCGCAGGUUCGAAUCCUGCUGAGUCCGCC --........(((((....-..)))))...............((((((((((((...(((((..((...))..))))).((((((..((....)).))))))....))).))))))))). ( -39.70) >consensus __UAGCAUCAACCAGUGGUGACCUGGUCUAAAAACACCAA_CACGCACUCAUAGCUCAGCUGGAUAGAGUACUCGGCUACGAACCGAGCGGUCGCAGGUUCGAAUCCUCCUGAGUGCGCC ................((((..............)))).....((((((((((((...))))....((.(.(((((.......))))).).))(((((.......))))))))))))).. (-35.40 = -32.49 + -2.91)
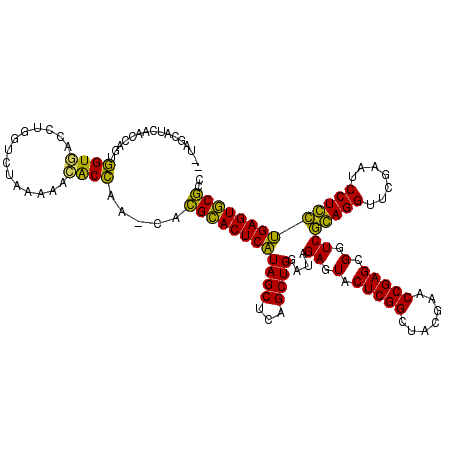
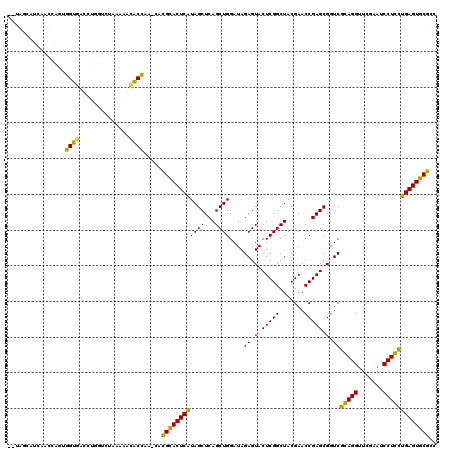
| Location | 991,615 – 991,731 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.37 |
| Mean single sequence MFE | -44.27 |
| Consensus MFE | -39.73 |
| Energy contribution | -36.73 |
| Covariance contribution | -2.99 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.926474 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.561 991615 116 - 6264404 GGCGCACUCAGGAGGAUUCGAACCUCCGACCGCUCGGUUCGUAGCCGAGUACUCUAUCCAGCUGAGCUAUGAGUGCGUG-UCGGUGCUUUU---UCCGGCCACCUGCCGGUAACGCUUGU .((((((((((((((.......)))))((..(((((((.....)))))))..))...............))))))))).-..((((.((..---.(((((.....))))).))))))... ( -46.40) >pfo.2515 1617599 116 + 6438405 GGUGCACUCGACAGGAUUCGAACCUGUGACCGCUCGGUUCGUAGCCGAGUACUCUAUCCAGCUGAGCUACGAGUGCAU--UUUGUGUUUUUAGACCAGAUCACAACUGGUUGAAGCCG-- (((((((((((((((.......)))))((..(((((((.....)))))))..))...............)))))))))--)..(.(((((..((((((.......)))))))))))).-- ( -40.90) >pf5.2742 1861359 116 + 7074893 GGUGCACUCGACAGGAUUCGAACCUGUGACCGCUCGGUUCGUAGCCGAGUACUCUAUCCAGCUGAGCUACGAGUGCAU--UUUGUGUUUUUAGACCAGACCACAACUGGUUGAAGCCA-- (((((((((((((((.......)))))((..(((((((.....)))))))..))...............)))))))))--).((.(((((..((((((.......)))))))))))))-- ( -41.00) >pel.2216 1732213 116 + 5888780 GGCGGACUCAGGAGGAUUCGAACCUCCGACCGCUCGGUUCGUAGCCGAGUACUCUAUCCAGCUGAGCUAUGAGUCCGUGG--GGUAGUUUUAGACCAGGUUACCACUGGUUGAUUGAA-- .((((((((((((((.......)))))((..(((((((.....)))))))..))...............)))))))))..--..(((((...((((((.......)))))))))))..-- ( -43.90) >pau.2472 1877673 116 + 6537648 GGCGCACUCAGGAGGAUUCGAACCUCCGACCGCUCGGUUCGUAGCCGAGUACUCUAUCCAGCUGAGCUAUGAGUGCGUG-UCGGUGCUUUU---UCCGGCCACCUGCCGGUAACGCUUGU .((((((((((((((.......)))))((..(((((((.....)))))))..))...............))))))))).-..((((.((..---.(((((.....))))).))))))... ( -46.40) >ppk.2577 1102295 117 + 6181863 GGCGGACUCAGCAGGAUUCGAACCUGCGACCGCUCGGUUCGUAGCCGAGUACUCUAUCCAGCUGAGCUAUGAGUCCGCAAUUGGCAGUUUUAGACCAGAU-ACCGCUGGUUGAUUGAA-- .((((((((((((((.......)))))((..(((((((.....)))))))..))...............)))))))))......(((((...((((((..-....)))))))))))..-- ( -47.00) >consensus GGCGCACUCAGCAGGAUUCGAACCUCCGACCGCUCGGUUCGUAGCCGAGUACUCUAUCCAGCUGAGCUAUGAGUGCGUG_UUGGUGCUUUUAGACCAGACCACCACUGGUUGAAGCAA__ .((((((((((((((.......)))))((..(((((((.....)))))))..))...............)))))))))....((((.((........)).))))................ (-39.73 = -36.73 + -2.99)

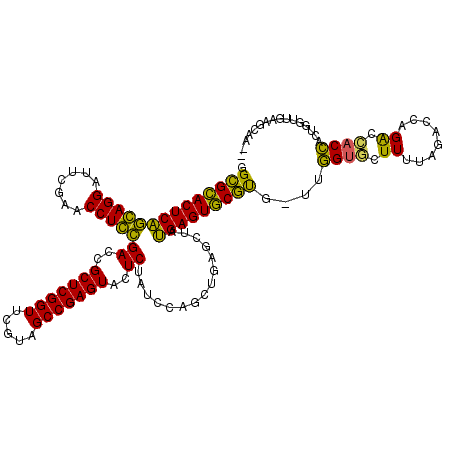
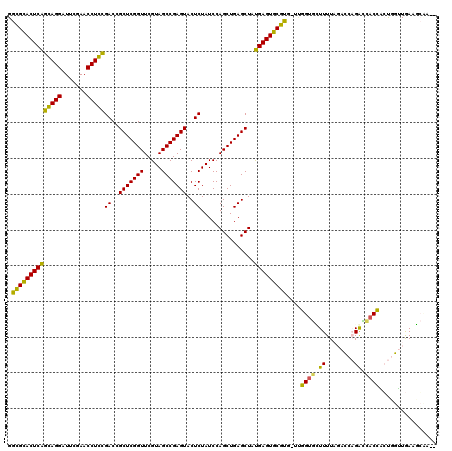
| Location | 991,617 – 991,733 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.89 |
| Mean single sequence MFE | -39.43 |
| Consensus MFE | -35.40 |
| Energy contribution | -32.49 |
| Covariance contribution | -2.91 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.706622 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
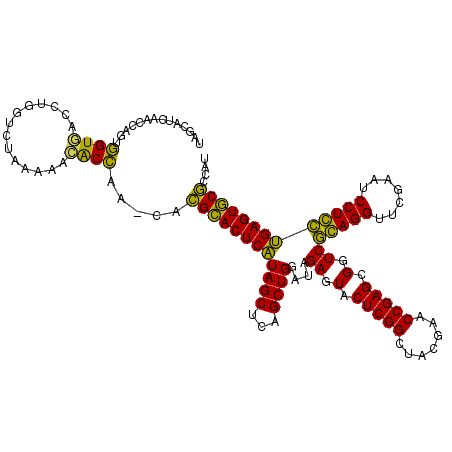
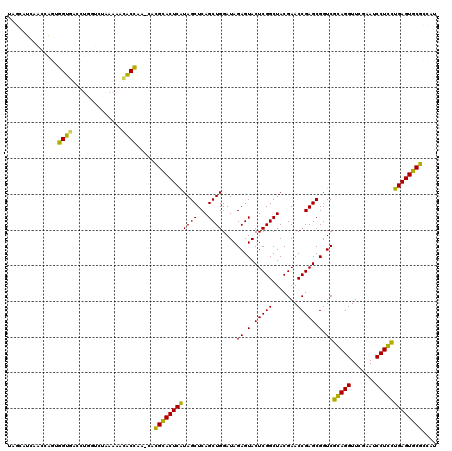
>pao.561 991617 116 + 6264404 AAGCGUUACCGGCAGGUGGCCGGA---AAAAGCACCGA-CACGCACUCAUAGCUCAGCUGGAUAGAGUACUCGGCUACGAACCGAGCGGUCGGAGGUUCGAAUCCUCCUGAGUGCGCCAU ..((....(((((.....))))).---....(((((((-(.(((((((.((((...))))....))))..((((.......))))))))))(((((.......))))).).))))))... ( -44.00) >pfo.2515 1617599 118 - 6438405 CGGCUUCAACCAGUUGUGAUCUGGUCUAAAAACACAAA--AUGCACUCGUAGCUCAGCUGGAUAGAGUACUCGGCUACGAACCGAGCGGUCACAGGUUCGAAUCCUGUCGAGUGCACCAU .((.....(((((.......))))).............--.(((((((((((...(((((..((...))..))))).((((((...........))))))....))).)))))))))).. ( -35.60) >pf5.2742 1861359 118 - 7074893 UGGCUUCAACCAGUUGUGGUCUGGUCUAAAAACACAAA--AUGCACUCGUAGCUCAGCUGGAUAGAGUACUCGGCUACGAACCGAGCGGUCACAGGUUCGAAUCCUGUCGAGUGCACCAU (((.....(((((.......))))).............--.(((((((((((...(((((..((...))..))))).((((((...........))))))....))).))))))))))). ( -35.80) >pel.2216 1732213 117 - 5888780 UUCAAUCAACCAGUGGUAACCUGGUCUAAAACUACC--CCACGGACUCAUAGCUCAGCUGGAUAGAGUACUCGGCUACGAACCGAGCGGUCGGAGGUUCGAAUCCUCCUGAGUCCGCCA- ........(((((.(....))))))...........--...((((((((((((...))))....((.(.(((((.......))))).).))(((((.......)))))))))))))...- ( -37.50) >pau.2472 1877675 116 - 6537648 AAGCGUUACCGGCAGGUGGCCGGA---AAAAGCACCGA-CACGCACUCAUAGCUCAGCUGGAUAGAGUACUCGGCUACGAACCGAGCGGUCGGAGGUUCGAAUCCUCCUGAGUGCGCCAU ..((....(((((.....))))).---....(((((((-(.(((((((.((((...))))....))))..((((.......))))))))))(((((.......))))).).))))))... ( -44.00) >ppk.2577 1102295 119 - 6181863 UUCAAUCAACCAGCGGU-AUCUGGUCUAAAACUGCCAAUUGCGGACUCAUAGCUCAGCUGGAUAGAGUACUCGGCUACGAACCGAGCGGUCGCAGGUUCGAAUCCUGCUGAGUCCGCCAC ........(((((....-..)))))...............((((((((((((...(((((..((...))..))))).((((((..((....)).))))))....))).)))))))))... ( -39.70) >consensus UAGCAUCAACCAGUGGUGACCUGGUCUAAAAACACCAA_CACGCACUCAUAGCUCAGCUGGAUAGAGUACUCGGCUACGAACCGAGCGGUCGCAGGUUCGAAUCCUCCUGAGUGCGCCAU ..............((((..............)))).....((((((((((((...))))....((.(.(((((.......))))).).))(((((.......))))))))))))).... (-35.40 = -32.49 + -2.91)

| Location | 991,617 – 991,733 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.89 |
| Mean single sequence MFE | -44.35 |
| Consensus MFE | -39.73 |
| Energy contribution | -36.73 |
| Covariance contribution | -2.99 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932765 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
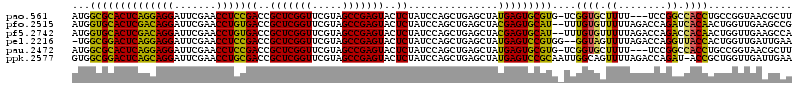
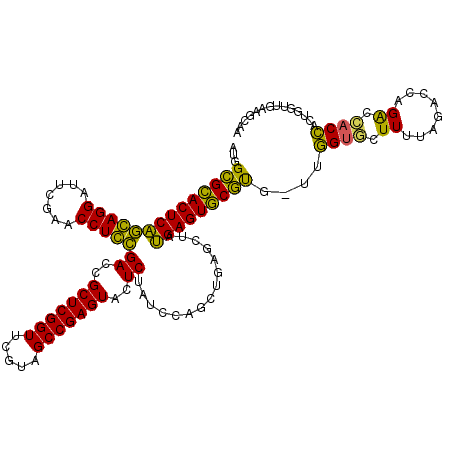
>pao.561 991617 116 - 6264404 AUGGCGCACUCAGGAGGAUUCGAACCUCCGACCGCUCGGUUCGUAGCCGAGUACUCUAUCCAGCUGAGCUAUGAGUGCGUG-UCGGUGCUUUU---UCCGGCCACCUGCCGGUAACGCUU ...((((((((((((((.......)))))((..(((((((.....)))))))..))...............))))))))).-..((((.((..---.(((((.....))))).)))))). ( -46.40) >pfo.2515 1617599 118 + 6438405 AUGGUGCACUCGACAGGAUUCGAACCUGUGACCGCUCGGUUCGUAGCCGAGUACUCUAUCCAGCUGAGCUACGAGUGCAU--UUUGUGUUUUUAGACCAGAUCACAACUGGUUGAAGCCG ..(((((((((((((((.......)))))((..(((((((.....)))))))..))...............)))))))))--)..(.(((((..((((((.......)))))))))))). ( -41.10) >pf5.2742 1861359 118 + 7074893 AUGGUGCACUCGACAGGAUUCGAACCUGUGACCGCUCGGUUCGUAGCCGAGUACUCUAUCCAGCUGAGCUACGAGUGCAU--UUUGUGUUUUUAGACCAGACCACAACUGGUUGAAGCCA ..(((((((((((((((.......)))))((..(((((((.....)))))))..))...............)))))))))--).((.(((((..((((((.......))))))))))))) ( -41.20) >pel.2216 1732213 117 + 5888780 -UGGCGGACUCAGGAGGAUUCGAACCUCCGACCGCUCGGUUCGUAGCCGAGUACUCUAUCCAGCUGAGCUAUGAGUCCGUGG--GGUAGUUUUAGACCAGGUUACCACUGGUUGAUUGAA -(.((((((((((((((.......)))))((..(((((((.....)))))))..))...............))))))))).)--..(((((...((((((.......))))))))))).. ( -44.00) >pau.2472 1877675 116 + 6537648 AUGGCGCACUCAGGAGGAUUCGAACCUCCGACCGCUCGGUUCGUAGCCGAGUACUCUAUCCAGCUGAGCUAUGAGUGCGUG-UCGGUGCUUUU---UCCGGCCACCUGCCGGUAACGCUU ...((((((((((((((.......)))))((..(((((((.....)))))))..))...............))))))))).-..((((.((..---.(((((.....))))).)))))). ( -46.40) >ppk.2577 1102295 119 + 6181863 GUGGCGGACUCAGCAGGAUUCGAACCUGCGACCGCUCGGUUCGUAGCCGAGUACUCUAUCCAGCUGAGCUAUGAGUCCGCAAUUGGCAGUUUUAGACCAGAU-ACCGCUGGUUGAUUGAA ...((((((((((((((.......)))))((..(((((((.....)))))))..))...............)))))))))......(((((...((((((..-....))))))))))).. ( -47.00) >consensus AUGGCGCACUCAGCAGGAUUCGAACCUCCGACCGCUCGGUUCGUAGCCGAGUACUCUAUCCAGCUGAGCUAUGAGUGCGUG_UUGGUGCUUUUAGACCAGACCACCACUGGUUGAAGCAA ...((((((((((((((.......)))))((..(((((((.....)))))))..))...............)))))))))....((((.((........)).)))).............. (-39.73 = -36.73 + -2.99)
Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:11:29 2007