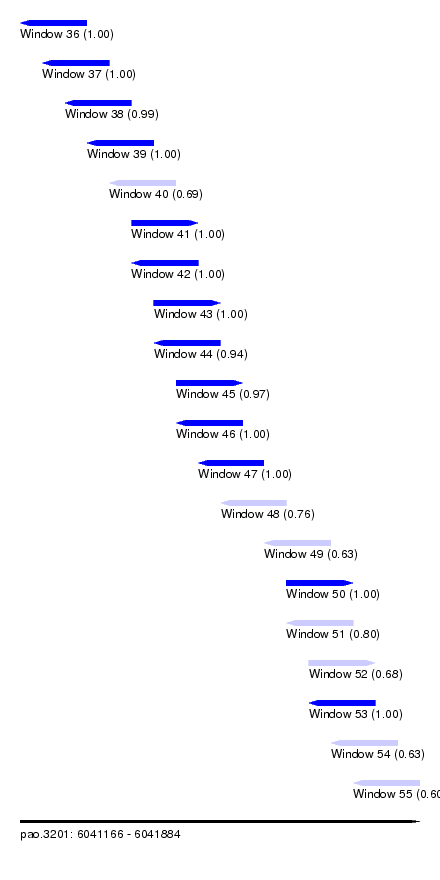
| Sequence ID | pao.3201 |
|---|---|
| Location | 6,041,166 – 6,041,884 |
| Length | 718 |
| Max. P | 0.999996 |

| Location | 6,041,166 – 6,041,286 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.16 |
| Mean single sequence MFE | -36.37 |
| Consensus MFE | -38.39 |
| Energy contribution | -35.12 |
| Covariance contribution | -3.27 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.52 |
| Structure conservation index | 1.06 |
| SVM decision value | 3.51 |
| SVM RNA-class probability | 0.999322 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
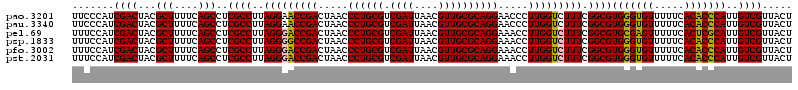
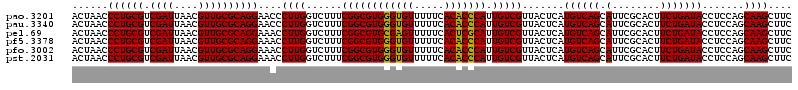
>pao.3201 6041166 120 - 6264404 GUUUAAGGUGGUAGGCUGAAAUCUUAGGUAAAUCCGGGGUUUCAAGGCCGAGAGCUGAUGACGAGUCGUCUUUUAGAUGACGAAGUGGUUGAUGCCAUGCUUCCAAGAAAAGCUUCUAAG (((...(((....((((((((((((.((.....))))))))))..))))....)))...)))..((((((.....))))))((((((((....)))))((((.......))))))).... ( -34.70) >pss.1919 3849032 120 - 6093698 GUUUAAGGUGGUAGGCUGAGAUCUUAGGUAAAUCCGGGAUCUUAAGGCCGAGAGCUGAUGACGAGUGUUCUUUUAGAACACGAAGUGGUUGAUGCCAUGCUUCCAAGAAAAGCUUCUAAG (((...(((....((((((((((((.((.....))))))))))..))))....)))...)))..((((((.....))))))((((((((....)))))((((.......))))))).... ( -36.40) >pel.69 5769698 119 + 5888780 GUUUAAGGUGGUAGGCUGAGAUCUUAGGCAAAUCCGGGAUCUCAAGGCCGAGAGCUGAUGACGAGUGCUC-AUUAGAGCGCGAAGUGGUUGAUGCCAUGCUUCCAAGAAAAGCUCCUAAG .....(((.(((.(((((((((((..((....))..)))))))..))))(..(((.........((((((-....))))))...(((((....))))))))..).......))))))... ( -40.60) >pf5.3378 6384323 120 - 7074893 GUUUAAGGUGGUAGGCUGGAAUCUUAGGUAAAUCCGGGGUUCUAAGGCCGAGAGCUGAUGACGAGUUACCUUUUAGGUGACGAAGUGGUUGAUGCCAUGCUUCCAAGAAAAGCUUCUAAG (((...(((....((((((((((((.((.....))))))))))..))))....)))...)))..((((((.....))))))((((((((....)))))((((.......))))))).... ( -35.60) >pfo.3002 5727944 119 - 6438405 GUUUAAGGUGGUAGGCUGAAAUCUUAGGUAAAUCCGGGGUUUCAAGGCCGAGAGCUGAUGACGAGUUGCC-UUUAGGCGACGAAGUGGUUGAUGCCAUGCUUCCAAGAAAAGCUUCUAAG (((...(((....((((((((((((.((.....))))))))))..))))....)))...)))..((((((-....))))))((((((((....)))))((((.......))))))).... ( -36.50) >pst.2031 3871033 120 - 6397126 GUUUAAGGUGGUAGGCUGAGAUCUUAGGUAAAUCCGGGAUCUUAAGGCCGAGAGCUGAUGACGAGUGUUCUUUUAGAACAUGAAGUGGUUGAUGCCAUGCUUCCAAGAAAAGCUUCUAAG .......((((((((((((((((((.((.....))))))))))..)))).......(((.((..((((((.....))))))...)).)))..))))))((((.......))))....... ( -34.40) >consensus GUUUAAGGUGGUAGGCUGAAAUCUUAGGUAAAUCCGGGAUCUCAAGGCCGAGAGCUGAUGACGAGUGGUCUUUUAGAACACGAAGUGGUUGAUGCCAUGCUUCCAAGAAAAGCUUCUAAG .......((((((((((((((((((.((.....))))))))))..)))).......(((.((..((((((.....))))))...)).)))..))))))((((.......))))....... (-38.39 = -35.12 + -3.27)

| Location | 6,041,206 – 6,041,326 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.55 |
| Mean single sequence MFE | -43.70 |
| Consensus MFE | -46.32 |
| Energy contribution | -43.13 |
| Covariance contribution | -3.19 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.73 |
| Structure conservation index | 1.06 |
| SVM decision value | 6.02 |
| SVM RNA-class probability | 0.999996 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.3201 6041206 120 - 6264404 GGACGGAGAAGGCUAGGCCAGCUUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGAAAUCUUAGGUAAAUCCGGGGUUUCAAGGCCGAGAGCUGAUGACGAGUCGUCUUUUAGAUGA ((((((..((.((((((....)))))).))..))))))..(((...(((....((((((((((((.((.....))))))))))..))))....)))...)))...(((((.....))))) ( -41.50) >pss.1919 3849072 120 - 6093698 GGACGGAGAAGGCUAGGCCAGCCUGGCGUUGGUUGUCCAGGUUUAAGGUGGUAGGCUGAGAUCUUAGGUAAAUCCGGGAUCUUAAGGCCGAGAGCUGAUGACGAGUGUUCUUUUAGAACA ((((((..((.((((((....)))))).))..))))))..(((...(((....((((((((((((.((.....))))))))))..))))....)))...)))...(((((.....))))) ( -45.70) >pel.69 5769738 119 + 5888780 GGACGGAGAAGGCUAGGCCAGCUUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGAGAUCUUAGGCAAAUCCGGGAUCUCAAGGCCGAGAGCUGAUGACGAGUGCUC-AUUAGAGCG ((((((..((.((((((....)))))).))..))))))..(((...(((....(((((((((((..((....))..)))))))..))))....)))...)))....((((-....)))). ( -46.50) >pf5.3378 6384363 120 - 7074893 GGACGGAGAAGGCUAGGCCAGCUUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGGAAUCUUAGGUAAAUCCGGGGUUCUAAGGCCGAGAGCUGAUGACGAGUUACCUUUUAGGUGA ((((((..((.((((((....)))))).))..))))))..(((...(((....((((((((((((.((.....))))))))))..))))....)))...)))...(((((.....))))) ( -42.40) >pfo.3002 5727984 119 - 6438405 GGACGGAGAAGGCUAGGCCAGCUUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGAAAUCUUAGGUAAAUCCGGGGUUUCAAGGCCGAGAGCUGAUGACGAGUUGCC-UUUAGGCGA ((((((..((.((((((....)))))).))..))))))..(((...(((....((((((((((((.((.....))))))))))..))))....)))...)))...(((((-....))))) ( -43.30) >pst.2031 3871073 120 - 6397126 GGACGGAGAAGGCUAGGCCAGCCUGGCGUUGGUAGUCCAGGUUUAAGGUGGUAGGCUGAGAUCUUAGGUAAAUCCGGGAUCUUAAGGCCGAGAGCUGAUGACGAGUGUUCUUUUAGAACA ((((....((.((((((....)))))).))....))))..(((...(((....((((((((((((.((.....))))))))))..))))....)))...)))...(((((.....))))) ( -42.80) >consensus GGACGGAGAAGGCUAGGCCAGCUUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGAAAUCUUAGGUAAAUCCGGGAUCUCAAGGCCGAGAGCUGAUGACGAGUGGUCUUUUAGAACA ((((((..((.((((((....)))))).))..))))))..(((...(((....((((((((((((.((.....))))))))))..))))....)))...)))...(((((.....))))) (-46.32 = -43.13 + -3.19)

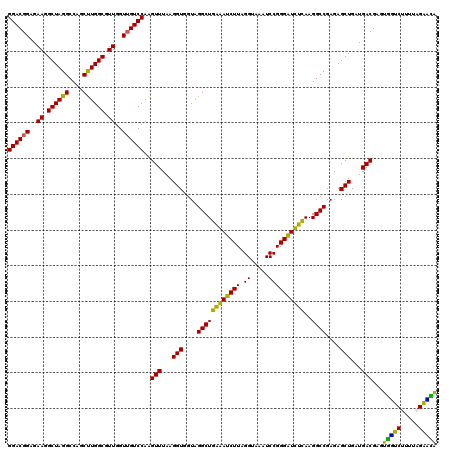
| Location | 6,041,246 – 6,041,366 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.28 |
| Mean single sequence MFE | -38.17 |
| Consensus MFE | -36.98 |
| Energy contribution | -36.65 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.990568 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
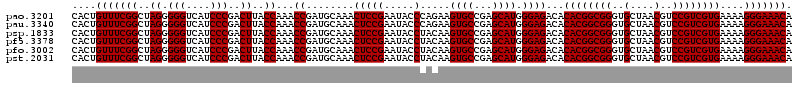
>pao.3201 6041246 120 - 6264404 ACAGGUUAAUAUUCCUGUACUUCUGGUUACUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCUUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGAAAUCUUAGGUAAAUCCGGGGU (((((........)))))..(((((((((((..(((....((((((..((.((((((....)))))).))..)))))).((((((......))))))...)))...)))))..)))))). ( -36.40) >pss.1919 3849112 120 - 6093698 ACAGGUUAAUAUUCCUGUACUUCUGGUUAUUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCCUGGCGUUGGUUGUCCAGGUUUAAGGUGGUAGGCUGAGAUCUUAGGUAAAUCCGGGAU (((((........)))))..((((((...((((...(((.((((((..((.((((((....)))))).))..)))))).((((((......))))))....)))...))))..)))))). ( -39.80) >pel.69 5769777 120 + 5888780 ACAGGUUAAUAUUCCUGUACUUCCAGUUAUUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCUUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGAGAUCUUAGGCAAAUCCGGGAU (((((........)))))..((((.((....))...(((.((((((..((.((((((....)))))).))..))))))..(((((((((((....))...)))))))))...))))))). ( -38.20) >psp.1833 3633116 120 - 5928787 ACAGGUUAAUAUUCCUGUACUUCUGGUUAUUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCCUGGCGUUGGUUGUCCAGGUUUAAGGUGGUAGGCUGAGAUCUUAGGUAAAUCCGGGAU (((((........)))))..((((((...((((...(((.((((((..((.((((((....)))))).))..)))))).((((((......))))))....)))...))))..)))))). ( -39.80) >pf5.3378 6384403 120 - 7074893 ACAGGUUAAUAUUCCUGUACUUCUAGUUAUUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCUUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGGAAUCUUAGGUAAAUCCGGGGU (((((........)))))..((((((((((..(..((((.((((((..((.((((((....)))))).))..))))))...))))..)..))).))))))).((..((.....))..)). ( -37.90) >pst.2031 3871113 120 - 6397126 ACAGGUUAAUAUUCCUGUACUUCUGGUUAUUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCCUGGCGUUGGUAGUCCAGGUUUAAGGUGGUAGGCUGAGAUCUUAGGUAAAUCCGGGAU (((((........)))))..((((((...((((...(((.((((....((.((((((....)))))).))....)))).((((((......))))))....)))...))))..)))))). ( -36.90) >consensus ACAGGUUAAUAUUCCUGUACUUCUGGUUAUUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCCUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGAGAUCUUAGGUAAAUCCGGGAU (((((........)))))..((((((...((((...(((.((((((..((.((((((....)))))).))..)))))).((((((......))))))....)))...))))..)))))). (-36.98 = -36.65 + -0.33)

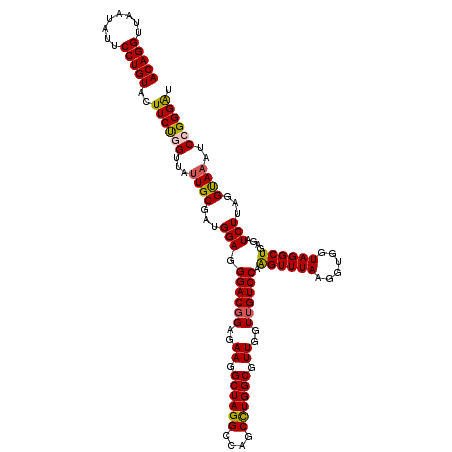
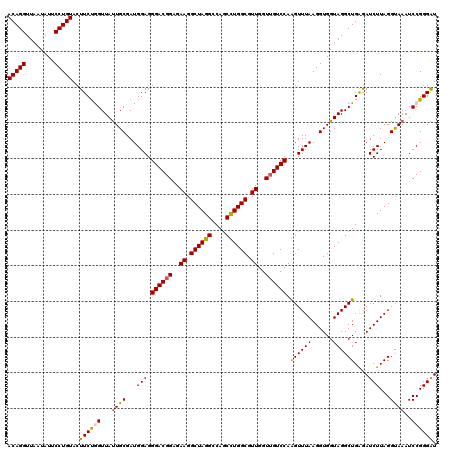
| Location | 6,041,286 – 6,041,406 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.50 |
| Mean single sequence MFE | -42.93 |
| Consensus MFE | -41.16 |
| Energy contribution | -40.47 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.96 |
| SVM decision value | 3.41 |
| SVM RNA-class probability | 0.999168 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.3201 6041286 120 - 6264404 CGGUUCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGGAAACAGGUUAAUAUUCCUGUACUUCUGGUUACUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCUUGGCGUUGGUUGUCCAA ...((((.........(((....)))(((((.(((.(((((((((........)))))..)))).))))))))...))))((((((..((.((((((....)))))).))..)))))).. ( -43.70) >pau.3340 6314205 120 - 6537648 CGGUUCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGGAAACAGGUUAAUAUUCCUGUACUUCUGGUUACUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCUUGGCGUUGGUUGUCCAA ...((((.........(((....)))(((((.(((.(((((((((........)))))..)))).))))))))...))))((((((..((.((((((....)))))).))..)))))).. ( -43.70) >pel.69 5769817 120 + 5888780 CGGUCCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUUCCAGUUAUUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCUUGGCGUUGGUUGUCCAA ..((((((....(...(((....)))(((((...(((((.(((((........)))))..)))))...))))))....))))))(((...(((((((((.....))).)))))).))).. ( -44.50) >psp.1833 3633156 120 - 5928787 CGGCCCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUUCUGGUUAUUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCCUGGCGUUGGUUGUCCAG .....((...(((.(.(((....))).).)))...))...(((((........))))).(((((.((....)).).))))((((((..((.((((((....)))))).))..)))))).. ( -42.20) >pf5.3378 6384443 120 - 7074893 CGGUCCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUUCUAGUUAUUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCUUGGCGUUGGUUGUCCAA ..((((((....(...(((....)))(((((...(((((.(((((........)))))..)))))...))))))....))))))(((...(((((((((.....))).)))))).))).. ( -42.20) >pst.2031 3871153 120 - 6397126 CGGUCCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUUCUGGUUAUUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCCUGGCGUUGGUAGUCCAG .(((..((((.((.((((((....((.(((((........(((((........))))).(((((.((....)).).))))..........))))).)))))))).)).))))..).)).. ( -41.30) >consensus CGGUCCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUUCUGGUUAUUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCUUGGCGUUGGUUGUCCAA ..((((((....(...(((....)))(((((...(((((.(((((........)))))..)))))...))))))....))))))(((...(((((((((.....))).)))))).))).. (-41.16 = -40.47 + -0.69)

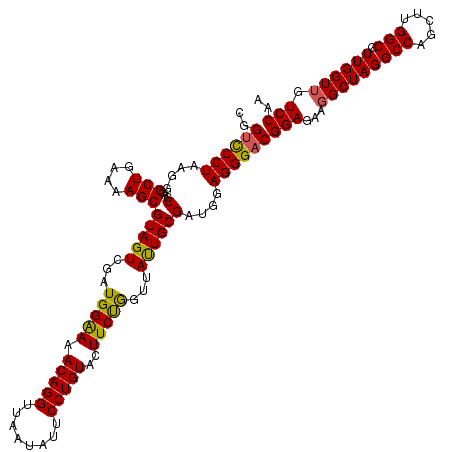
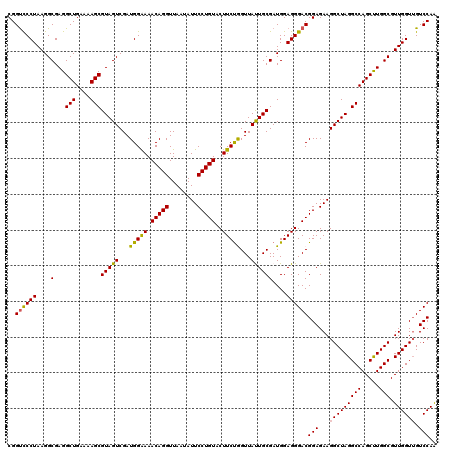
| Location | 6,041,326 – 6,041,446 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -37.05 |
| Consensus MFE | -36.63 |
| Energy contribution | -35.47 |
| Covariance contribution | -1.16 |
| Combinations/Pair | 1.16 |
| Mean z-score | -0.72 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.692405 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.3201 6041326 120 - 6264404 CCAAGGGUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUUCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGGAAACAGGUUAAUAUUCCUGUACUUCUGGUUACUGCGAUGGAG (((......((((((...((.....)).))))))(((..(((((..(......)..)))))..)))(((((.(((.(((((((((........)))))..)))).))))))))..))).. ( -38.40) >pau.3340 6314245 120 - 6537648 CCAAGGGUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUUCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGGAAACAGGUUAAUAUUCCUGUACUUCUGGUUACUGCGAUGGAG (((......((((((...((.....)).))))))(((..(((((..(......)..)))))..)))(((((.(((.(((((((((........)))))..)))).))))))))..))).. ( -38.40) >pel.69 5769857 120 + 5888780 CCAAGGUUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUCCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUUCCAGUUAUUGCGAUGGAG (((.....(((((((...((.....)).)))))))((((((.(((.....)))..))))))....((((((...(((((.(((((........)))))..)))))...)))))).))).. ( -37.80) >psp.1833 3633196 120 - 5928787 CCAAGGUUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGCCCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUUCUGGUUAUUGCGAUGGAG (((.....(((((((...((.....)).)))))))((((((.(((.....)))..))))))....((((((...(((((.(((((........)))))..)))))...)))))).))).. ( -36.90) >pf5.3378 6384483 120 - 7074893 CCAAGGUUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUCCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUUCUAGUUAUUGCGAUGGAG (((.....(((((((...((.....)).)))))))((((((.(((.....)))..))))))....((((((...(((((.(((((........)))))..)))))...)))))).))).. ( -35.50) >pst.2031 3871193 120 - 6397126 CCAAGGUUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUCCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUUCUGGUUAUUGCGAUGGAG ........(((..(((((.....((((((((((((.........))))...))...(((....)))...))))))((((.(((((........)))))..)))).....)))))..))). ( -35.30) >consensus CCAAGGUUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUCCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUUCUGGUUAUUGCGAUGGAG (((.....(((((((...((.....)).)))))))((((((.(((.....)))..))))))....((((((...(((((.(((((........)))))..)))))...)))))).))).. (-36.63 = -35.47 + -1.16)
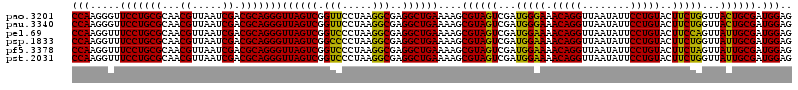
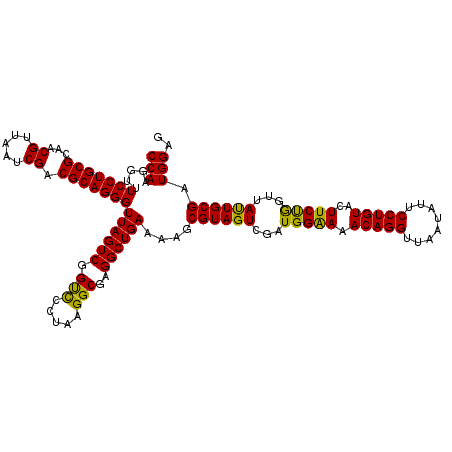
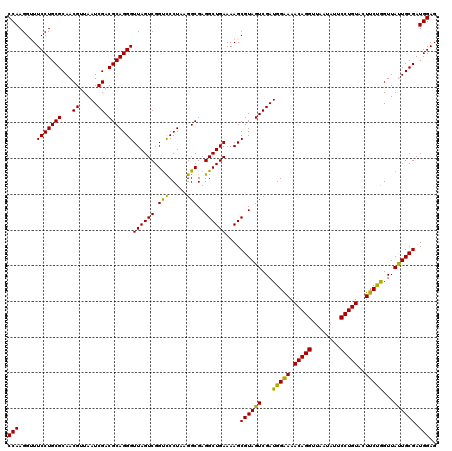
| Location | 6,041,366 – 6,041,486 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.28 |
| Mean single sequence MFE | -43.05 |
| Consensus MFE | -42.56 |
| Energy contribution | -42.20 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.66 |
| Structure conservation index | 0.99 |
| SVM decision value | 5.80 |
| SVM RNA-class probability | 0.999994 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.3201 6041366 120 + 6264404 UUCCCAUCGACUACGCUUUUCAGCCUCGCCUUAGGAACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAACCCUUGGUCUUUCGGCGUGGGUGUUUUUCACACCCAUUGUCGUUACU .......((((...(((....)))..((((..((((.((((.....((((((.((((....)))))))))).....))))))))..))))(((((((.....)))))))..))))..... ( -41.40) >pau.3340 6314285 120 + 6537648 UUCCCAUCGACUACGCUUUUCAGCCUCGCCUUAGGAACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAACCCUUGGUCUUUCGGCGUGGGUGUUUUUCACACCCAUUGUCGUUACU .......((((...(((....)))..((((..((((.((((.....((((((.((((....)))))))))).....))))))))..))))(((((((.....)))))))..))))..... ( -41.40) >pel.69 5769897 120 - 5888780 UUUCCAUCGACUACGCUUUUCAGCCUCGCCUUAGGGACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGCGAGUUUUUCACUCGCAUUGUCGUUACU .......((((...(((....)))..((((..(((((((((.....((((((.((((....)))))))))).....))))))))).))))(((((((.....)))))))..))))..... ( -43.30) >psp.1833 3633236 120 + 5928787 UUUCCAUCGACUACGCUUUUCAGCCUCGCCUUAGGGGCCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGGGUGUUUUUCACACCCAUUGUCGUUACU .......((((.(((((.....(((((......))))).((((((.((((((.((((....)))))))))).....))))))....)))))((((((.....))))))...))))..... ( -44.40) >pfo.3002 5728143 120 + 6438405 UUUCCAUCGACUACGCUUUUCAGCCUCGCCUUAGGGACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGGGUGUUUUUCACACCCAUUGUCGUUACU .......((((...(((....)))..((((..(((((((((.....((((((.((((....)))))))))).....))))))))).))))(((((((.....)))))))..))))..... ( -43.90) >pst.2031 3871233 120 + 6397126 UUUCCAUCGACUACGCUUUUCAGCCUCGCCUUAGGGACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGGGUGUUUUUCACACCCAUUGUCGUUACU .......((((...(((....)))..((((..(((((((((.....((((((.((((....)))))))))).....))))))))).))))(((((((.....)))))))..))))..... ( -43.90) >consensus UUUCCAUCGACUACGCUUUUCAGCCUCGCCUUAGGGACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGGGUGUUUUUCACACCCAUUGUCGUUACU .......((((...(((....)))..((((..(((((((((.....((((((.((((....)))))))))).....))))))))).))))(((((((.....)))))))..))))..... (-42.56 = -42.20 + -0.36)
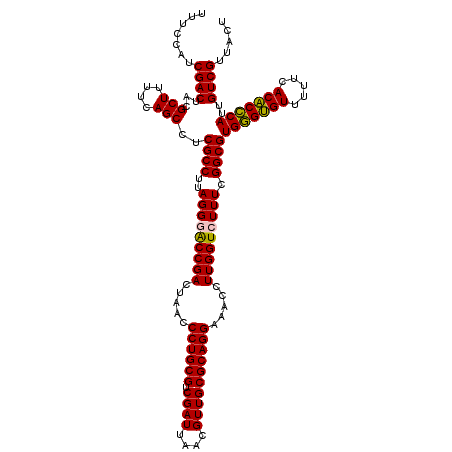
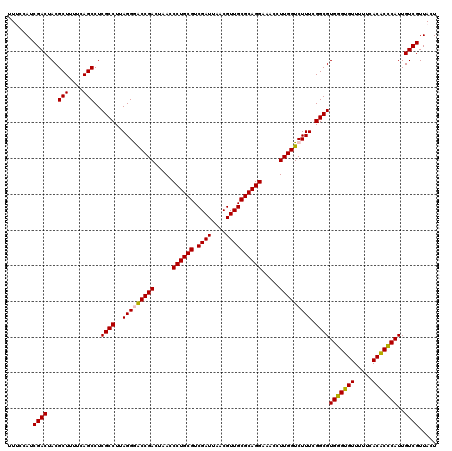
| Location | 6,041,366 – 6,041,486 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.28 |
| Mean single sequence MFE | -44.62 |
| Consensus MFE | -44.94 |
| Energy contribution | -44.33 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.48 |
| Structure conservation index | 1.01 |
| SVM decision value | 6.04 |
| SVM RNA-class probability | 0.999996 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
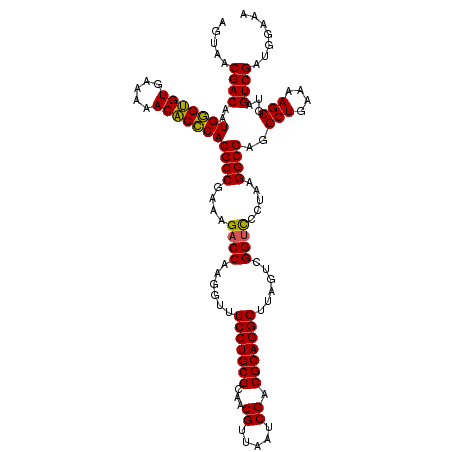
>pao.3201 6041366 120 - 6264404 AGUAACGACAAUGGGUGUGAAAAACACCCACGCCGAAAGACCAAGGGUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUUCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGGAA .....((((..(((((((.....)))))))((((....((((......(((((((...((.....)).)))))))......)))).....))))..(((....)))...))))....... ( -44.00) >pau.3340 6314285 120 - 6537648 AGUAACGACAAUGGGUGUGAAAAACACCCACGCCGAAAGACCAAGGGUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUUCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGGAA .....((((..(((((((.....)))))))((((....((((......(((((((...((.....)).)))))))......)))).....))))..(((....)))...))))....... ( -44.00) >pel.69 5769897 120 + 5888780 AGUAACGACAAUGCGAGUGAAAAACUCGCACGCCGAAAGACCAAGGUUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUCCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGAAA .....((((..(((((((.....)))))))((((....((((......(((((((...((.....)).)))))))......)))).....))))..(((....)))...))))....... ( -45.20) >psp.1833 3633236 120 - 5928787 AGUAACGACAAUGGGUGUGAAAAACACCCACGCCGAAAGACCAAGGUUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGCCCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGAAA .....((((...((((((.....))))))(((((....).........(((((((...((.....)).)))))))((((((.(((.....)))..))))))...)))).))))....... ( -42.90) >pfo.3002 5728143 120 - 6438405 AGUAACGACAAUGGGUGUGAAAAACACCCACGCCGAAAGACCAAGGUUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUCCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGAAA .....((((..(((((((.....)))))))((((....((((......(((((((...((.....)).)))))))......)))).....))))..(((....)))...))))....... ( -45.80) >pst.2031 3871233 120 - 6397126 AGUAACGACAAUGGGUGUGAAAAACACCCACGCCGAAAGACCAAGGUUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUCCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGAAA .....((((..(((((((.....)))))))((((....((((......(((((((...((.....)).)))))))......)))).....))))..(((....)))...))))....... ( -45.80) >consensus AGUAACGACAAUGGGUGUGAAAAACACCCACGCCGAAAGACCAAGGUUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUCCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGAAA .....((((..(((((((.....)))))))((((....((((......(((((((...((.....)).)))))))......)))).....))))..(((....)))...))))....... (-44.94 = -44.33 + -0.61)

| Location | 6,041,406 – 6,041,526 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.44 |
| Mean single sequence MFE | -34.70 |
| Consensus MFE | -35.25 |
| Energy contribution | -34.70 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.96 |
| Structure conservation index | 1.02 |
| SVM decision value | 4.21 |
| SVM RNA-class probability | 0.999839 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.3201 6041406 120 + 6264404 ACUAACCCUGCGUCGAUUAACGUUGCGCAGGAACCCUUGGUCUUUCGGCGUGGGUGUUUUUCACACCCAUUGUCGUUACUCAUGUCAGCAUUCGCACUUCUGAUACCUCCAGCAAGCUUC ......((((((.((((....))))))))))....((((......((((((((((((.....))))))).))))).......((((((.(........))))))).......)))).... ( -34.80) >pau.3340 6314325 120 + 6537648 ACUAACCCUGCGUCGAUUAACGUUGCGCAGGAACCCUUGGUCUUUCGGCGUGGGUGUUUUUCACACCCAUUGUCGUUACUCAUGUCAGCAUUCGCACUUCUGAUACCUCCAGCAAGCUUC ......((((((.((((....))))))))))....((((......((((((((((((.....))))))).))))).......((((((.(........))))))).......)))).... ( -34.80) >pel.69 5769937 120 - 5888780 ACUAACCCUGCGUCGAUUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGCGAGUUUUUCACUCGCAUUGUCGUUACUCAUGUCAGCAUUCGCACUUCUGAUACCUCCAGCAAGCUUC ......((((((.((((....))))))))))....((((......((((((((((((.....))))))).))))).......((((((.(........))))))).......)))).... ( -34.20) >pf5.3378 6384563 120 + 7074893 ACUAACCCUGCGUCGAUUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGGGUGUUUUUCACACCCAUUGUCGUUACUCAUGUCAGCAUUCGCACUUCUGAUACCUCCAGCAAGCUUC ......((((((.((((....))))))))))....((((......((((((((((((.....))))))).))))).......((((((.(........))))))).......)))).... ( -34.80) >pfo.3002 5728183 120 + 6438405 ACUAACCCUGCGUCGAUUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGGGUGUUUUUCACACCCAUUGUCGUUACUCAUGUCAGCAUUCGCACUUCUGAUACCUCCAGCAAGCUUC ......((((((.((((....))))))))))....((((......((((((((((((.....))))))).))))).......((((((.(........))))))).......)))).... ( -34.80) >pst.2031 3871273 120 + 6397126 ACUAACCCUGCGUCGAUUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGGGUGUUUUUCACACCCAUUGUCGUUACUCAUGUCAGCAUUCGCACUUCUGAUACCUCCAGCAAGCUUC ......((((((.((((....))))))))))....((((......((((((((((((.....))))))).))))).......((((((.(........))))))).......)))).... ( -34.80) >consensus ACUAACCCUGCGUCGAUUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGGGUGUUUUUCACACCCAUUGUCGUUACUCAUGUCAGCAUUCGCACUUCUGAUACCUCCAGCAAGCUUC ......((((((.((((....))))))))))....((((......((((((((((((.....))))))).))))).......((((((.(........))))))).......)))).... (-35.25 = -34.70 + -0.55)
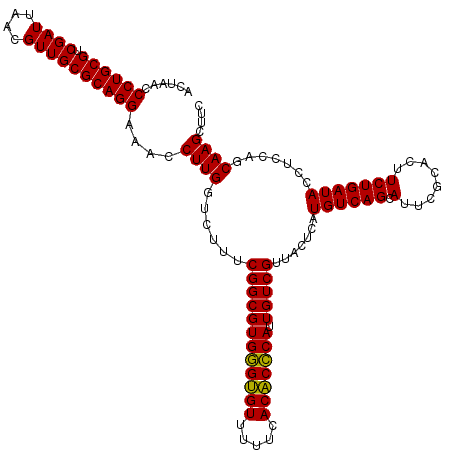
| Location | 6,041,406 – 6,041,526 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.44 |
| Mean single sequence MFE | -35.33 |
| Consensus MFE | -34.95 |
| Energy contribution | -34.40 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.05 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.944662 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.3201 6041406 120 - 6264404 GAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUGAAAAACACCCACGCCGAAAGACCAAGGGUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGU .((.(((((((..(((((..(.....)..)))))..))(((.(((((....(((((((.....)))))))((((....)....(((...))))))...))))).)))..))))).))... ( -34.50) >pau.3340 6314325 120 - 6537648 GAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUGAAAAACACCCACGCCGAAAGACCAAGGGUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGU .((.(((((((..(((((..(.....)..)))))..))(((.(((((....(((((((.....)))))))((((....)....(((...))))))...))))).)))..))))).))... ( -34.50) >pel.69 5769937 120 + 5888780 GAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGCGAGUGAAAAACUCGCACGCCGAAAGACCAAGGUUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGU .((.(((((((..(((((..(.....)..)))))..))(((.(((((....(((((((.....))))))).((((..((((....))))..)).))..))))).)))..))))).))... ( -35.30) >pf5.3378 6384563 120 - 7074893 GAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUGAAAAACACCCACGCCGAAAGACCAAGGUUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGU .((.(((((((..(((((..(.....)..)))))..))(((.(((((....(((((((.....))))))).((((..((((....))))..)).))..))))).)))..))))).))... ( -35.90) >pfo.3002 5728183 120 - 6438405 GAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUGAAAAACACCCACGCCGAAAGACCAAGGUUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGU .((.(((((((..(((((..(.....)..)))))..))(((.(((((....(((((((.....))))))).((((..((((....))))..)).))..))))).)))..))))).))... ( -35.90) >pst.2031 3871273 120 - 6397126 GAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUGAAAAACACCCACGCCGAAAGACCAAGGUUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGU .((.(((((((..(((((..(.....)..)))))..))(((.(((((....(((((((.....))))))).((((..((((....))))..)).))..))))).)))..))))).))... ( -35.90) >consensus GAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUGAAAAACACCCACGCCGAAAGACCAAGGUUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGU .((.(((((((..(((((..(.....)..)))))..))(((.(((((....(((((((.....)))))))((((....)....(((...))))))...))))).)))..))))).))... (-34.95 = -34.40 + -0.55)

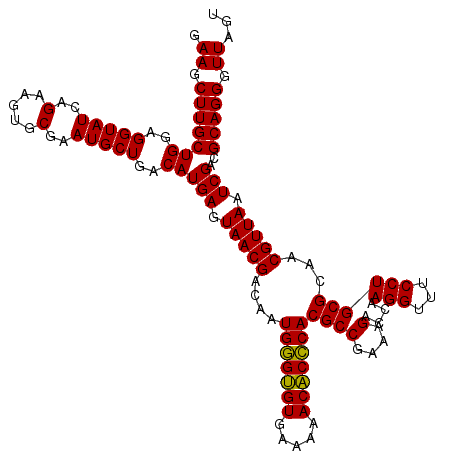

| Location | 6,041,446 – 6,041,566 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -23.70 |
| Consensus MFE | -24.25 |
| Energy contribution | -23.70 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.02 |
| Structure conservation index | 1.02 |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.971211 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.3201 6041446 120 + 6264404 UCUUUCGGCGUGGGUGUUUUUCACACCCAUUGUCGUUACUCAUGUCAGCAUUCGCACUUCUGAUACCUCCAGCAAGCUUCUCAACUCACCUUCACAGGCUUACAGAACGCUCCUCUACCG .....((((((((((((.....))))))).))))).......((((((.(........))))))).........(((((((.......(((....))).....)))).)))......... ( -23.80) >pel.69 5769977 120 - 5888780 UCUUUCGGCGUGCGAGUUUUUCACUCGCAUUGUCGUUACUCAUGUCAGCAUUCGCACUUCUGAUACCUCCAGCAAGCUUCUCAACUCACCUUCACAGGCUUACAGAACGCUCCUCUACCG .....((((((((((((.....))))))).))))).......((((((.(........))))))).........(((((((.......(((....))).....)))).)))......... ( -23.20) >psp.1833 3633316 120 + 5928787 UCUUUCGGCGUGGGUGUUUUUCACACCCAUUGUCGUUACUCAUGUCAGCAUUCGCACUUCUGAUACCUCCAGCAAGCUUCUCAACUCACCUUCACAGGCUUACAGAACGCUCCUCUACCG .....((((((((((((.....))))))).))))).......((((((.(........))))))).........(((((((.......(((....))).....)))).)))......... ( -23.80) >pf5.3378 6384603 120 + 7074893 UCUUUCGGCGUGGGUGUUUUUCACACCCAUUGUCGUUACUCAUGUCAGCAUUCGCACUUCUGAUACCUCCAGCAAGCUUCUCAACUCACCUUCACAGGCUUACAGAACGCUCCUCUACCG .....((((((((((((.....))))))).))))).......((((((.(........))))))).........(((((((.......(((....))).....)))).)))......... ( -23.80) >pfo.3002 5728223 120 + 6438405 UCUUUCGGCGUGGGUGUUUUUCACACCCAUUGUCGUUACUCAUGUCAGCAUUCGCACUUCUGAUACCUCCAGCAAGCUUCUCAACUCACCUUCACAGGCUUACAGAACGCUCCUCUACCG .....((((((((((((.....))))))).))))).......((((((.(........))))))).........(((((((.......(((....))).....)))).)))......... ( -23.80) >pst.2031 3871313 120 + 6397126 UCUUUCGGCGUGGGUGUUUUUCACACCCAUUGUCGUUACUCAUGUCAGCAUUCGCACUUCUGAUACCUCCAGCAAGCUUCUCAACUCACCUUCACAGGCUUACAGAACGCUCCUCUACCG .....((((((((((((.....))))))).))))).......((((((.(........))))))).........(((((((.......(((....))).....)))).)))......... ( -23.80) >consensus UCUUUCGGCGUGGGUGUUUUUCACACCCAUUGUCGUUACUCAUGUCAGCAUUCGCACUUCUGAUACCUCCAGCAAGCUUCUCAACUCACCUUCACAGGCUUACAGAACGCUCCUCUACCG .....((((((((((((.....))))))).))))).......((((((.(........))))))).........(((((((.......(((....))).....)))).)))......... (-24.25 = -23.70 + -0.55)

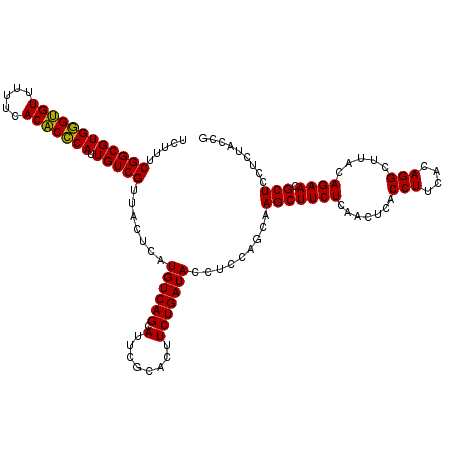
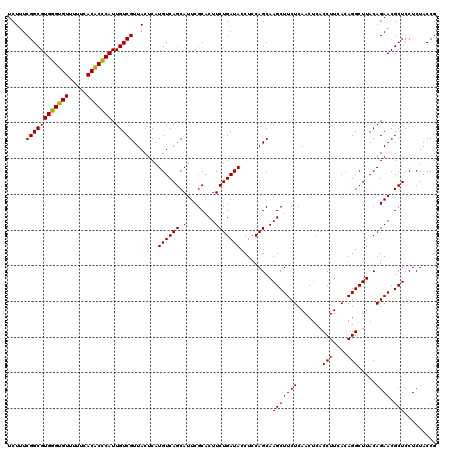
| Location | 6,041,446 – 6,041,566 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -38.10 |
| Consensus MFE | -38.65 |
| Energy contribution | -38.10 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.64 |
| Structure conservation index | 1.01 |
| SVM decision value | 5.27 |
| SVM RNA-class probability | 0.999981 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.3201 6041446 120 - 6264404 CGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUGAAAAACACCCACGCCGAAAGA ((((......((((((((...((((.(...((..((((....))))..))).))))..))))).)))...((((......)))).......(((((((.....))))))).))))..... ( -38.20) >pel.69 5769977 120 + 5888780 CGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGCGAGUGAAAAACUCGCACGCCGAAAGA ((((......((((((((...((((.(...((..((((....))))..))).))))..))))).)))...((((......)))).......(((((((.....))))))).))))..... ( -37.60) >psp.1833 3633316 120 - 5928787 CGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUGAAAAACACCCACGCCGAAAGA ((((......((((((((...((((.(...((..((((....))))..))).))))..))))).)))...((((......)))).......(((((((.....))))))).))))..... ( -38.20) >pf5.3378 6384603 120 - 7074893 CGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUGAAAAACACCCACGCCGAAAGA ((((......((((((((...((((.(...((..((((....))))..))).))))..))))).)))...((((......)))).......(((((((.....))))))).))))..... ( -38.20) >pfo.3002 5728223 120 - 6438405 CGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUGAAAAACACCCACGCCGAAAGA ((((......((((((((...((((.(...((..((((....))))..))).))))..))))).)))...((((......)))).......(((((((.....))))))).))))..... ( -38.20) >pst.2031 3871313 120 - 6397126 CGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUGAAAAACACCCACGCCGAAAGA ((((......((((((((...((((.(...((..((((....))))..))).))))..))))).)))...((((......)))).......(((((((.....))))))).))))..... ( -38.20) >consensus CGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUGAAAAACACCCACGCCGAAAGA ((((......((((((((...((((.(...((..((((....))))..))).))))..))))).)))...((((......)))).......(((((((.....))))))).))))..... (-38.65 = -38.10 + -0.55)

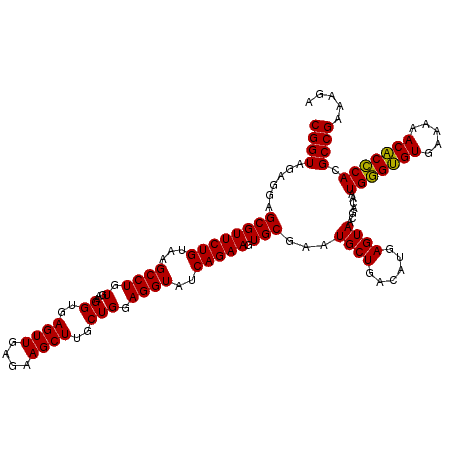
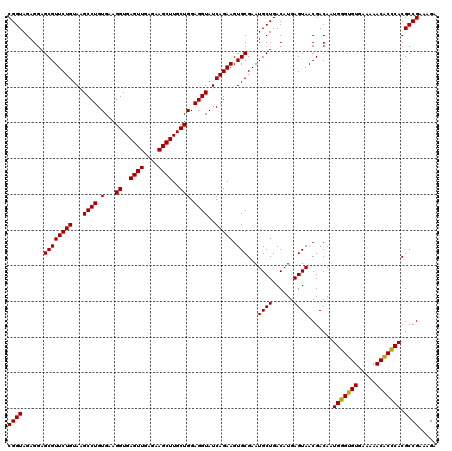
| Location | 6,041,486 – 6,041,604 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.53 |
| Mean single sequence MFE | -40.22 |
| Consensus MFE | -37.46 |
| Energy contribution | -36.52 |
| Covariance contribution | -0.94 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.92 |
| SVM RNA-class probability | 0.997719 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.3201 6041486 118 - 6264404 AACCACACACCGAAGCUGCGGGUGUCACGU--AAGUGACGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUG .(((.....(((......)))(((((((..--..))))))))))......((((((((...((((.(...((..((((....))))..))).))))..))))).)))............. ( -40.50) >pau.3340 6314405 118 - 6537648 AACCACACACCGAAGCUGCGGGUGUCACGC--AAGUGACGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUG .(((.....(((......)))(((((((..--..))))))))))......((((((((...((((.(...((..((((....))))..))).))))..))))).)))............. ( -41.50) >pss.1919 3849352 120 - 6093698 AACCAUGCACCGAAGCUACGGGUAUCAUCUUAGGAUGAUGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUG ...((.(((((....((((..((((((((....)))))))).)))).)).((((((((...((((.(...((..((((....))))..))).))))..))))).)))...)))))..... ( -39.60) >psp.1833 3633356 120 - 5928787 AACCAUGCACCGAAGCUACGGGUAUCAUCUUUUGAUGAUGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUG ...((.(((((....((((..((((((((....)))))))).)))).)).((((((((...((((.(...((..((((....))))..))).))))..))))).)))...)))))..... ( -39.80) >pfo.3002 5728263 118 - 6438405 AACCAUACACCGAAGCUACGGGUAUCACGC--AAGUGAUGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUG ..((.(((.(((......)))(((((((..--..))))))).)))..)).((((((((...((((.(...((..((((....))))..))).))))..))))).)))............. ( -40.10) >pst.2031 3871353 120 - 6397126 AACCAUGCACCGAAGCUACGGGUAUCAUCUUUUGAUGAUGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUG ...((.(((((....((((..((((((((....)))))))).)))).)).((((((((...((((.(...((..((((....))))..))).))))..))))).)))...)))))..... ( -39.80) >consensus AACCAUACACCGAAGCUACGGGUAUCACCU__AAAUGAUGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUG .(((.....(((......)))(((((((......))))))))))......((((((((...((((.(...((..((((....))))..))).))))..))))).)))............. (-37.46 = -36.52 + -0.94)
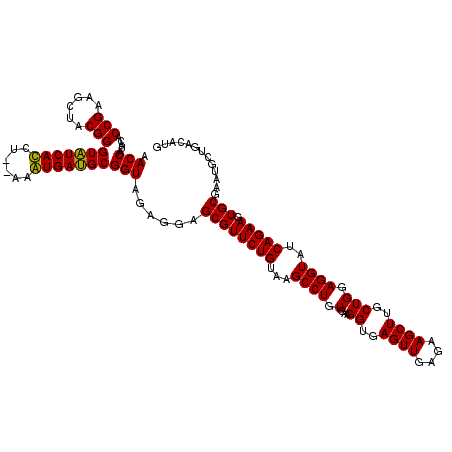
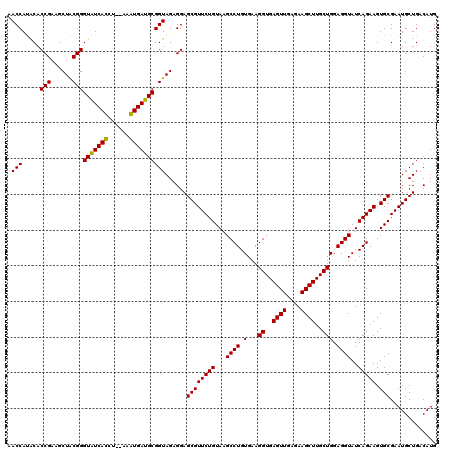
| Location | 6,041,526 – 6,041,644 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.53 |
| Mean single sequence MFE | -41.32 |
| Consensus MFE | -39.56 |
| Energy contribution | -38.62 |
| Covariance contribution | -0.94 |
| Combinations/Pair | 1.05 |
| Mean z-score | -0.96 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.757464 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.3201 6041526 118 - 6264404 CUAGUCGAGUCGGCCUGCGCGGAAGAUGUAACGGGGCUCAAACCACACACCGAAGCUGCGGGUGUCACGU--AAGUGACGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGA ....((((.(((.(((.(((((.....((.(((((((((..(((.....(((......)))(((((((..--..))))))))))....))))..)))))..))))))).)))))).)))) ( -42.60) >pau.3340 6314445 118 - 6537648 CUAGUCGAGUCGGCCUGCGCGGAAGAUGUAACGGGGCUCAAACCACACACCGAAGCUGCGGGUGUCACGC--AAGUGACGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGA ....((((.(((.(((.(((((.....((.(((((((((..(((.....(((......)))(((((((..--..))))))))))....))))..)))))..))))))).)))))).)))) ( -43.60) >pss.1919 3849392 120 - 6093698 CUAGUCGAGUCGGCCUGCGCGGAAGAUGUAACGGGGCUCAAACCAUGCACCGAAGCUACGGGUAUCAUCUUAGGAUGAUGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGA ....((((.(((.(((.(((((.....((.(((((((((..(((.....(((......)))((((((((....)))))))))))....))))..)))))..))))))).)))))).)))) ( -39.80) >psp.1833 3633396 120 - 5928787 CUAGUCGAGUCGGCCUGCGCGGAAGAUGUAACGGGGCUCAAACCAUGCACCGAAGCUACGGGUAUCAUCUUUUGAUGAUGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGA ....((((.(((.(((.(((((.....((.(((((((((..(((.....(((......)))((((((((....)))))))))))....))))..)))))..))))))).)))))).)))) ( -40.00) >pfo.3002 5728303 118 - 6438405 CUAGUCGAGUCGGCCUGCGCGGAAGAUGUAACGGGGCUCAAACCAUACACCGAAGCUACGGGUAUCACGC--AAGUGAUGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGA ....((((.(((.(((.(((((.....((.(((((((((..(((.....(((......)))(((((((..--..))))))))))....))))..)))))..))))))).)))))).)))) ( -41.90) >pst.2031 3871393 120 - 6397126 CUAGUCGAGUCGGCCUGCGCGGAAGAUGUAACGGGGCUCAAACCAUGCACCGAAGCUACGGGUAUCAUCUUUUGAUGAUGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGA ....((((.(((.(((.(((((.....((.(((((((((..(((.....(((......)))((((((((....)))))))))))....))))..)))))..))))))).)))))).)))) ( -40.00) >consensus CUAGUCGAGUCGGCCUGCGCGGAAGAUGUAACGGGGCUCAAACCAUACACCGAAGCUACGGGUAUCACCU__AAAUGAUGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGA ....((((.(((.(((.(((((.....((.(((((((((..(((.....(((......)))(((((((......))))))))))....))))..)))))..))))))).)))))).)))) (-39.56 = -38.62 + -0.94)

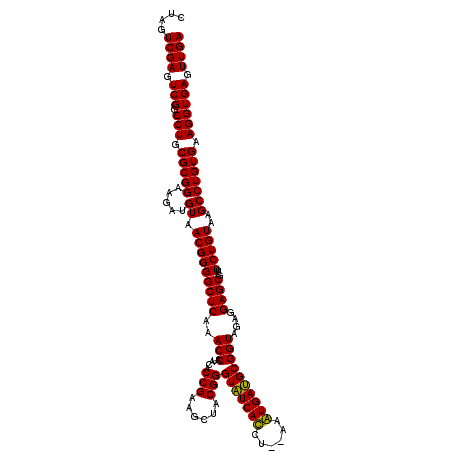
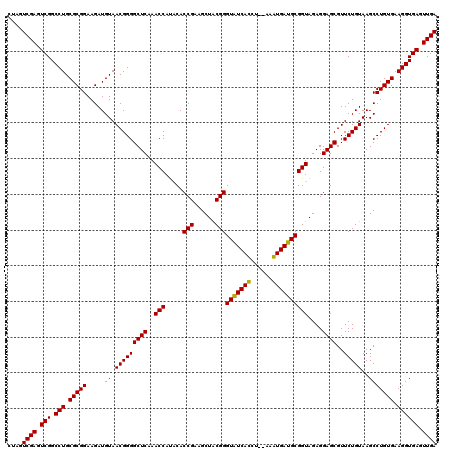
| Location | 6,041,604 – 6,041,724 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.56 |
| Mean single sequence MFE | -36.80 |
| Consensus MFE | -35.80 |
| Energy contribution | -36.13 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.78 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.628156 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.3201 6041604 120 - 6264404 GGUAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCAGCCACCCUUUAAAGAAAGCGUAAUAGCUCACUAGUCGAGUCGGCCUGCGCGGAAGAUGUAACGGGGCUCA (((...((..(..((..(((((.(((.....(((((.(((((.......))))).)))))..((..(((......)))..)).))))))))..))..).(....)......))..))).. ( -35.40) >pau.3340 6314523 120 - 6537648 GGUAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCAGCCACCCUUUAAAGAAAGCGUAAUAGCUCACUAGUCGAGUCGGCCUGCGCGGAAGAUGUAACGGGGCUCA (((...((..(..((..(((((.(((.....(((((.(((((.......))))).)))))..((..(((......)))..)).))))))))..))..).(....)......))..))).. ( -35.40) >psp.1833 3633476 120 - 5928787 GGGAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCAGCCACCCUUUAAAGAAAGCGUAAUAGCUCACUAGUCGAGUCGGCCUGCGCGGAAGAUGUAACGGGGCUCA (((...((..(..((..(((((.(((.....(((((.(((((.......))))).)))))..((..(((......)))..)).))))))))..))..).(....)......))...))). ( -37.50) >pf5.3378 6384761 120 - 7074893 GGGAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCAGCCACCCUUUAAAGAAAGCGUAAUAGCUCACUAGUCGAGUCGGCCUGCGCGGAAGAUGUAACGGGGCUCA (((...((..(..((..(((((.(((.....(((((.(((((.......))))).)))))..((..(((......)))..)).))))))))..))..).(....)......))...))). ( -37.50) >pfo.3002 5728381 120 - 6438405 GGGAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCAGCCACCCUUUAAAGAAAGCGUAAUAGCUCACUAGUCGAGUCGGCCUGCGCGGAAGAUGUAACGGGGCUCA (((...((..(..((..(((((.(((.....(((((.(((((.......))))).)))))..((..(((......)))..)).))))))))..))..).(....)......))...))). ( -37.50) >pst.2031 3871473 120 - 6397126 GGGAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCAGCCACCCUUUAAAGAAAGCGUAAUAGCUCACUAGUCGAGUCGGCCUGCGCGGAAGAUGUAACGGGGCUCA (((...((..(..((..(((((.(((.....(((((.(((((.......))))).)))))..((..(((......)))..)).))))))))..))..).(....)......))...))). ( -37.50) >consensus GGGAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCAGCCACCCUUUAAAGAAAGCGUAAUAGCUCACUAGUCGAGUCGGCCUGCGCGGAAGAUGUAACGGGGCUCA (((...((..(..((..(((((.(((.....(((((.(((((.......))))).)))))..((..(((......)))..)).))))))))..))..).(....)......))...))). (-35.80 = -36.13 + 0.33)

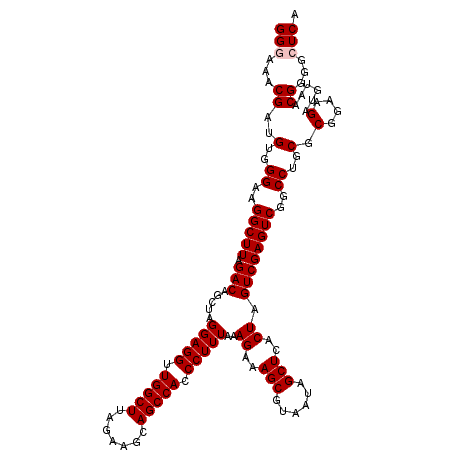
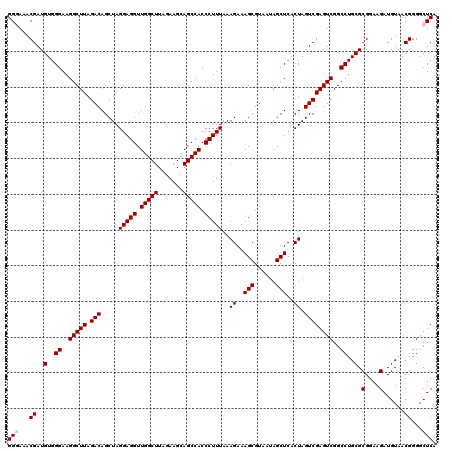
| Location | 6,041,644 – 6,041,764 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.22 |
| Mean single sequence MFE | -32.98 |
| Consensus MFE | -32.48 |
| Energy contribution | -32.04 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.91 |
| SVM RNA-class probability | 0.997702 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.3201 6041644 120 + 6264404 UGAGCUAUUACGCUUUCUUUAAAGGGUGGCUGCUUCUAAGCCAACCUCCUAGCUGUCUAAGCCUUCCCACAUCGUUUACCACUUAACCACAACUUUGGGACCUUAGCUGGCGGUCUGGGU .((((......)))).......(((.(((((.......))))).)))((((((((((..(((..(((((....(((........)))........))))).....)))))))).))))). ( -31.30) >pau.3340 6314563 120 + 6537648 UGAGCUAUUACGCUUUCUUUAAAGGGUGGCUGCUUCUAAGCCAACCUCCUAGCUGUCUAAGCCUUCCCACAUCGUUUACCACUUAACCACAACUUUGGGACCUUAGCUGGCGGUCUGGGU .((((......)))).......(((.(((((.......))))).)))((((((((((..(((..(((((....(((........)))........))))).....)))))))).))))). ( -31.30) >psp.1833 3633516 120 + 5928787 UGAGCUAUUACGCUUUCUUUAAAGGGUGGCUGCUUCUAAGCCAACCUCCUAGCUGUCUAAGCCUUCCCACAUCGUUUCCCACUUAACCAUGACUUUGGGACCUUAGCUGACGGUCUGGGU .((((......)))).......(((.(((((.......))))).)))((((((((((..(((..((((((((.(((........))).)))....))))).....)))))))).))))). ( -35.30) >pf5.3378 6384801 120 + 7074893 UGAGCUAUUACGCUUUCUUUAAAGGGUGGCUGCUUCUAAGCCAACCUCCUAGCUGUCUAAGCCUUCCCACAUCGUUUCCCACUUAACCAUAACUUUGGGACCUUAGCUGACGGUCUGGGU .((((......)))).......(((.(((((.......))))).)))((((((((((..(((..(((((....(((..............)))..))))).....)))))))).))))). ( -32.34) >pfo.3002 5728421 120 + 6438405 UGAGCUAUUACGCUUUCUUUAAAGGGUGGCUGCUUCUAAGCCAACCUCCUAGCUGUCUAAGCCUUCCCACAUCGUUUCCCACUUAACCAUAACUUUGGGACCUUAGCUGACGGUCUGGGU .((((......)))).......(((.(((((.......))))).)))((((((((((..(((..(((((....(((..............)))..))))).....)))))))).))))). ( -32.34) >pst.2031 3871513 120 + 6397126 UGAGCUAUUACGCUUUCUUUAAAGGGUGGCUGCUUCUAAGCCAACCUCCUAGCUGUCUAAGCCUUCCCACAUCGUUUCCCACUUAACCAUGACUUUGGGACCUUAGCUGACGGUCUGGGU .((((......)))).......(((.(((((.......))))).)))((((((((((..(((..((((((((.(((........))).)))....))))).....)))))))).))))). ( -35.30) >consensus UGAGCUAUUACGCUUUCUUUAAAGGGUGGCUGCUUCUAAGCCAACCUCCUAGCUGUCUAAGCCUUCCCACAUCGUUUCCCACUUAACCAUAACUUUGGGACCUUAGCUGACGGUCUGGGU .((((......)))).......(((.(((((.......))))).)))((((((((((..(((..(((((....(((..............)))..))))).....)))))))).))))). (-32.48 = -32.04 + -0.44)
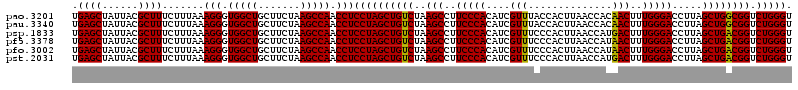
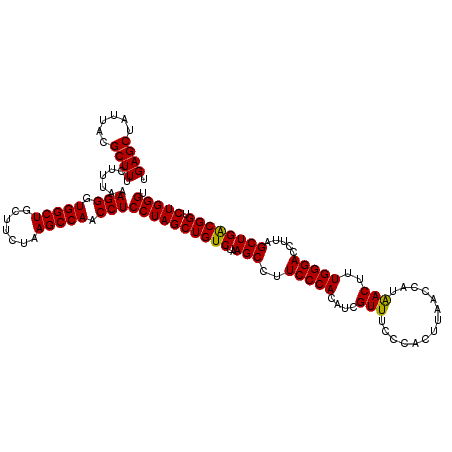
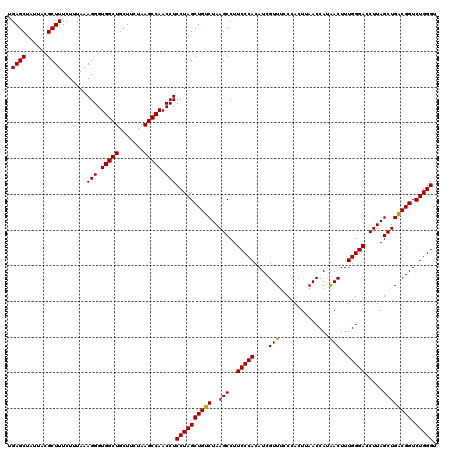
| Location | 6,041,644 – 6,041,764 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.22 |
| Mean single sequence MFE | -33.40 |
| Consensus MFE | -33.24 |
| Energy contribution | -33.13 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -0.78 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.795767 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.3201 6041644 120 - 6264404 ACCCAGACCGCCAGCUAAGGUCCCAAAGUUGUGGUUAAGUGGUAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCAGCCACCCUUUAAAGAAAGCGUAAUAGCUCA .((((((((((.((((..(....)..))))))))))..((.....))....))))..((((...((.(((.(((((.(((((.......))))).)))))......)))))...)))).. ( -33.40) >pau.3340 6314563 120 - 6537648 ACCCAGACCGCCAGCUAAGGUCCCAAAGUUGUGGUUAAGUGGUAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCAGCCACCCUUUAAAGAAAGCGUAAUAGCUCA .((((((((((.((((..(....)..))))))))))..((.....))....))))..((((...((.(((.(((((.(((((.......))))).)))))......)))))...)))).. ( -33.40) >psp.1833 3633516 120 - 5928787 ACCCAGACCGUCAGCUAAGGUCCCAAAGUCAUGGUUAAGUGGGAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCAGCCACCCUUUAAAGAAAGCGUAAUAGCUCA .((((((((((..(((..(....)..))).))))))..((.(....).)).))))..((((...((.(((.(((((.(((((.......))))).)))))......)))))...)))).. ( -32.50) >pf5.3378 6384801 120 - 7074893 ACCCAGACCGUCAGCUAAGGUCCCAAAGUUAUGGUUAAGUGGGAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCAGCCACCCUUUAAAGAAAGCGUAAUAGCUCA .((((((((((.((((..(....)..))))))))))..((.(....).)).))))..((((...((.(((.(((((.(((((.......))))).)))))......)))))...)))).. ( -34.30) >pfo.3002 5728421 120 - 6438405 ACCCAGACCGUCAGCUAAGGUCCCAAAGUUAUGGUUAAGUGGGAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCAGCCACCCUUUAAAGAAAGCGUAAUAGCUCA .((((((((((.((((..(....)..))))))))))..((.(....).)).))))..((((...((.(((.(((((.(((((.......))))).)))))......)))))...)))).. ( -34.30) >pst.2031 3871513 120 - 6397126 ACCCAGACCGUCAGCUAAGGUCCCAAAGUCAUGGUUAAGUGGGAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCAGCCACCCUUUAAAGAAAGCGUAAUAGCUCA .((((((((((..(((..(....)..))).))))))..((.(....).)).))))..((((...((.(((.(((((.(((((.......))))).)))))......)))))...)))).. ( -32.50) >consensus ACCCAGACCGUCAGCUAAGGUCCCAAAGUUAUGGUUAAGUGGGAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCAGCCACCCUUUAAAGAAAGCGUAAUAGCUCA .((((((((((.((((..(....)..))))))))))..((.(....).)).))))..((((...((.(((.(((((.(((((.......))))).)))))......)))))...)))).. (-33.24 = -33.13 + -0.11)

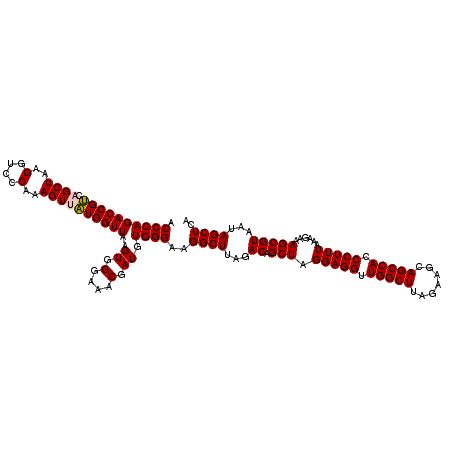
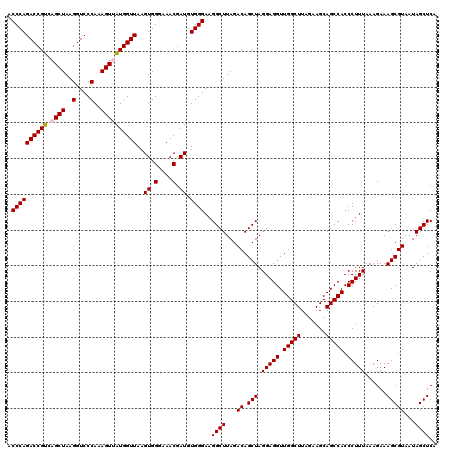
| Location | 6,041,684 – 6,041,804 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.22 |
| Mean single sequence MFE | -30.08 |
| Consensus MFE | -29.58 |
| Energy contribution | -29.14 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -0.73 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.677668 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.3201 6041684 120 + 6264404 CCAACCUCCUAGCUGUCUAAGCCUUCCCACAUCGUUUACCACUUAACCACAACUUUGGGACCUUAGCUGGCGGUCUGGGUUGUUUCCCUUUUCACGACGGACGUUAGCACCCGCCGUGUG ..(((..((((((((((..(((..(((((....(((........)))........))))).....)))))))).)))))..)))........((((.(((..(....)..))).)))).. ( -28.40) >pau.3340 6314603 120 + 6537648 CCAACCUCCUAGCUGUCUAAGCCUUCCCACAUCGUUUACCACUUAACCACAACUUUGGGACCUUAGCUGGCGGUCUGGGUUGUUUCCCUUUUCACGACGGACGUUAGCACCCGCCGUGUG ..(((..((((((((((..(((..(((((....(((........)))........))))).....)))))))).)))))..)))........((((.(((..(....)..))).)))).. ( -28.40) >psp.1833 3633556 120 + 5928787 CCAACCUCCUAGCUGUCUAAGCCUUCCCACAUCGUUUCCCACUUAACCAUGACUUUGGGACCUUAGCUGACGGUCUGGGUUGUUUCCCUUUUCACGACGGACGUUAGCACCCGCCGUGUG ..(((..((((((((((..(((..((((((((.(((........))).)))....))))).....)))))))).)))))..)))........((((.(((..(....)..))).)))).. ( -32.40) >pf5.3378 6384841 120 + 7074893 CCAACCUCCUAGCUGUCUAAGCCUUCCCACAUCGUUUCCCACUUAACCAUAACUUUGGGACCUUAGCUGACGGUCUGGGUUGUUUCCCUUUUCACGACGGACGUUAGCACCCGCCGUGUG ..(((..((((((((((..(((..(((((....(((..............)))..))))).....)))))))).)))))..)))........((((.(((..(....)..))).)))).. ( -29.44) >pfo.3002 5728461 120 + 6438405 CCAACCUCCUAGCUGUCUAAGCCUUCCCACAUCGUUUCCCACUUAACCAUAACUUUGGGACCUUAGCUGACGGUCUGGGUUGUUUCCCUUUUCACGACGGACGUUAGCACCCGCCGUGUG ..(((..((((((((((..(((..(((((....(((..............)))..))))).....)))))))).)))))..)))........((((.(((..(....)..))).)))).. ( -29.44) >pst.2031 3871553 120 + 6397126 CCAACCUCCUAGCUGUCUAAGCCUUCCCACAUCGUUUCCCACUUAACCAUGACUUUGGGACCUUAGCUGACGGUCUGGGUUGUUUCCCUUUUCACGACGGACGUUAGCACCCGCCGUGUG ..(((..((((((((((..(((..((((((((.(((........))).)))....))))).....)))))))).)))))..)))........((((.(((..(....)..))).)))).. ( -32.40) >consensus CCAACCUCCUAGCUGUCUAAGCCUUCCCACAUCGUUUCCCACUUAACCAUAACUUUGGGACCUUAGCUGACGGUCUGGGUUGUUUCCCUUUUCACGACGGACGUUAGCACCCGCCGUGUG ..(((..((((((((((..(((..(((((....(((..............)))..))))).....)))))))).)))))..)))........((((.(((..(....)..))).)))).. (-29.58 = -29.14 + -0.44)


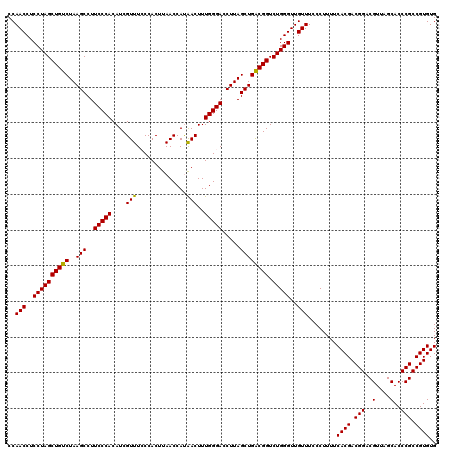
| Location | 6,041,684 – 6,041,804 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.22 |
| Mean single sequence MFE | -40.15 |
| Consensus MFE | -39.26 |
| Energy contribution | -38.60 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.98 |
| SVM decision value | 3.00 |
| SVM RNA-class probability | 0.998088 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.3201 6041684 120 - 6264404 CACACGGCGGGUGCUAACGUCCGUCGUGAAAAGGGAAACAACCCAGACCGCCAGCUAAGGUCCCAAAGUUGUGGUUAAGUGGUAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGG ..((((((((..(....)..))))))))....(((......))).((((.(((((((((.(((((..(((((.............))))).)))))...)))....)))).)).)))).. ( -41.62) >pau.3340 6314603 120 - 6537648 CACACGGCGGGUGCUAACGUCCGUCGUGAAAAGGGAAACAACCCAGACCGCCAGCUAAGGUCCCAAAGUUGUGGUUAAGUGGUAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGG ..((((((((..(....)..))))))))....(((......))).((((.(((((((((.(((((..(((((.............))))).)))))...)))....)))).)).)))).. ( -41.62) >psp.1833 3633556 120 - 5928787 CACACGGCGGGUGCUAACGUCCGUCGUGAAAAGGGAAACAACCCAGACCGUCAGCUAAGGUCCCAAAGUCAUGGUUAAGUGGGAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGG ..((((((((..(....)..))))))))....(((......))).((((..((((((((.(((((....(((.(((........))).))))))))...)))....)))).)..)))).. ( -39.90) >pf5.3378 6384841 120 - 7074893 CACACGGCGGGUGCUAACGUCCGUCGUGAAAAGGGAAACAACCCAGACCGUCAGCUAAGGUCCCAAAGUUAUGGUUAAGUGGGAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGG ..((((((((..(....)..))))))))....(((......))).((((..((((((((.(((((.............((.(....).)).)))))...)))....)))).)..)))).. ( -38.94) >pfo.3002 5728461 120 - 6438405 CACACGGCGGGUGCUAACGUCCGUCGUGAAAAGGGAAACAACCCAGACCGUCAGCUAAGGUCCCAAAGUUAUGGUUAAGUGGGAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGG ..((((((((..(....)..))))))))....(((......))).((((..((((((((.(((((.............((.(....).)).)))))...)))....)))).)..)))).. ( -38.94) >pst.2031 3871553 120 - 6397126 CACACGGCGGGUGCUAACGUCCGUCGUGAAAAGGGAAACAACCCAGACCGUCAGCUAAGGUCCCAAAGUCAUGGUUAAGUGGGAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGG ..((((((((..(....)..))))))))....(((......))).((((..((((((((.(((((....(((.(((........))).))))))))...)))....)))).)..)))).. ( -39.90) >consensus CACACGGCGGGUGCUAACGUCCGUCGUGAAAAGGGAAACAACCCAGACCGUCAGCUAAGGUCCCAAAGUUAUGGUUAAGUGGGAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGG ..((((((((..(....)..))))))))....(((......))).((((.(((((((((.(((((....(((.(((........))).))))))))...)))....)))).)).)))).. (-39.26 = -38.60 + -0.66)

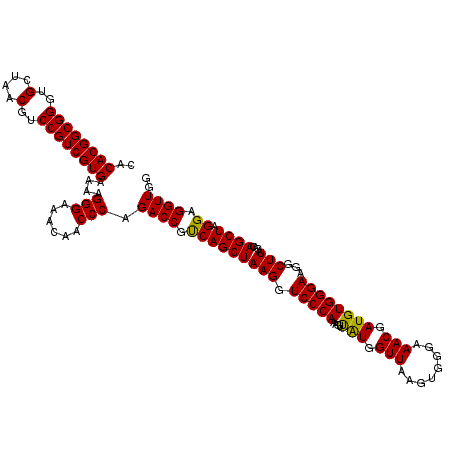
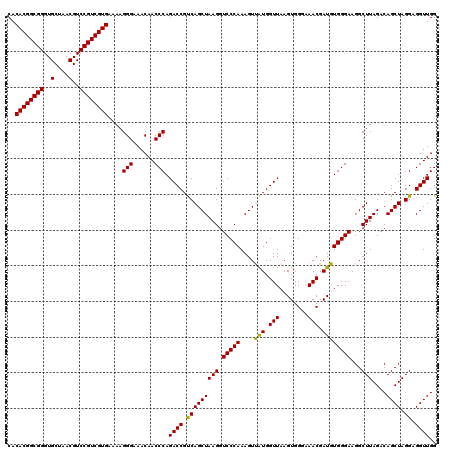
| Location | 6,041,724 – 6,041,844 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.78 |
| Mean single sequence MFE | -34.50 |
| Consensus MFE | -33.78 |
| Energy contribution | -33.67 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -0.72 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.631940 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.3201 6041724 120 - 6264404 AUGCAAACUCCGAAUACCCAGAAGUGCCGAGCAUGGGAGACACACGGCGGGUGCUAACGUCCGUCGUGAAAAGGGAAACAACCCAGACCGCCAGCUAAGGUCCCAAAGUUGUGGUUAAGU .......(((((......)....((((...)))).))))...((((((((..(....)..))))))))....(((......))).((((((.((((..(....)..)))))))))).... ( -36.40) >pau.3340 6314643 120 - 6537648 AUGCAAACUCCGAAUACCCAGAAGUGCCGAGCAUGGGAGACACACGGCGGGUGCUAACGUCCGUCGUGAAAAGGGAAACAACCCAGACCGCCAGCUAAGGUCCCAAAGUUGUGGUUAAGU .......(((((......)....((((...)))).))))...((((((((..(....)..))))))))....(((......))).((((((.((((..(....)..)))))))))).... ( -36.40) >psp.1833 3633596 120 - 5928787 AUGCAAACUCCGAAUACCUACAAGUGCCGAGCAUGGGAGACACACGGCGGGUGCUAACGUCCGUCGUGAAAAGGGAAACAACCCAGACCGUCAGCUAAGGUCCCAAAGUCAUGGUUAAGU .........(((....(((....((((...))))))).(((.((((((((..(....)..))))))))....(((......))).((((.........)))).....))).)))...... ( -33.20) >pf5.3378 6384881 120 - 7074893 AUGCAAACUCCGAAUACCUACAAGUGCCGAGCAUGGGAGACACACGGCGGGUGCUAACGUCCGUCGUGAAAAGGGAAACAACCCAGACCGUCAGCUAAGGUCCCAAAGUUAUGGUUAAGU .......(((((........)..((((...)))).))))...((((((((..(....)..))))))))....(((......))).((((((.((((..(....)..)))))))))).... ( -33.90) >pfo.3002 5728501 120 - 6438405 AUGCAAACUCCGAAUACCUACAAGUGCCGAGCAUGGGAGACACACGGCGGGUGCUAACGUCCGUCGUGAAAAGGGAAACAACCCAGACCGUCAGCUAAGGUCCCAAAGUUAUGGUUAAGU .......(((((........)..((((...)))).))))...((((((((..(....)..))))))))....(((......))).((((((.((((..(....)..)))))))))).... ( -33.90) >pst.2031 3871593 120 - 6397126 AUGCAAACUCCGAAUACCUACAAGUGCCGAGCAUGGGAGACACACGGCGGGUGCUAACGUCCGUCGUGAAAAGGGAAACAACCCAGACCGUCAGCUAAGGUCCCAAAGUCAUGGUUAAGU .........(((....(((....((((...))))))).(((.((((((((..(....)..))))))))....(((......))).((((.........)))).....))).)))...... ( -33.20) >consensus AUGCAAACUCCGAAUACCUACAAGUGCCGAGCAUGGGAGACACACGGCGGGUGCUAACGUCCGUCGUGAAAAGGGAAACAACCCAGACCGUCAGCUAAGGUCCCAAAGUUAUGGUUAAGU .......(((((.....).....((((...)))).))))...((((((((..(....)..))))))))....(((......))).((((((.((((..(....)..)))))))))).... (-33.78 = -33.67 + -0.11)

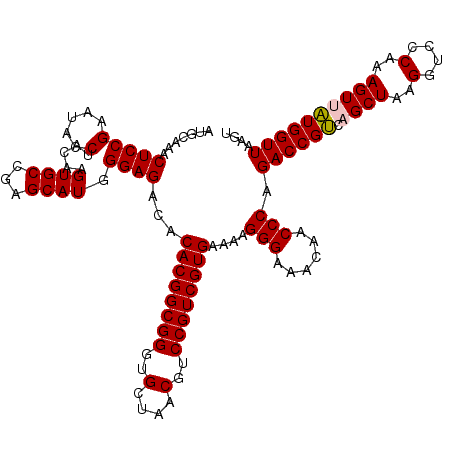
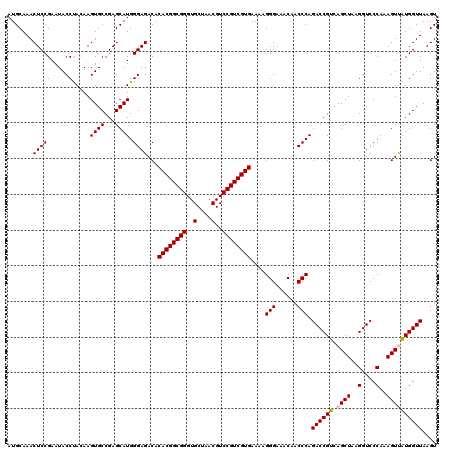
| Location | 6,041,764 – 6,041,884 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.11 |
| Mean single sequence MFE | -37.63 |
| Consensus MFE | -37.07 |
| Energy contribution | -37.07 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.69 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.596001 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.3201 6041764 120 - 6264404 CACUGUUUCGGCUAGGGGGUCAUCCCGACUUACCAAACCGAUGCAAACUCCGAAUACCCAGAAGUGCCGAGCAUGGGAGACACACGGCGGGUGCUAACGUCCGUCGUGAAAAGGGAAACA ...(((.((((...(((((((.....))))).))...)))).)))...(((.....((((...((.....)).)))).....((((((((..(....)..)))))))).....))).... ( -38.30) >pau.3340 6314683 120 - 6537648 CACUGUUUCGGCUAGGGGGUCAUCCCGACUUACCAAACCGAUGCAAACUCCGAAUACCCAGAAGUGCCGAGCAUGGGAGACACACGGCGGGUGCUAACGUCCGUCGUGAAAAGGGAAACA ...(((.((((...(((((((.....))))).))...)))).)))...(((.....((((...((.....)).)))).....((((((((..(....)..)))))))).....))).... ( -38.30) >psp.1833 3633636 120 - 5928787 CACUGUUUCGGCUAGGGGGUCAUCCCGACUUACCAAACCGAUGCAAACUCCGAAUACCUACAAGUGCCGAGCAUGGGAGACACACGGCGGGUGCUAACGUCCGUCGUGAAAAGGGAAACA ....(((((((..((.(((....)))..))..))...((........(((((........)..((((...)))).))))...((((((((..(....)..))))))))....))))))). ( -37.30) >pf5.3378 6384921 120 - 7074893 CACUGUUUCGGCUAGGGGGUCAUCCCGACUUACCAAACCGAUGCAAACUCCGAAUACCUACAAGUGCCGAGCAUGGGAGACACACGGCGGGUGCUAACGUCCGUCGUGAAAAGGGAAACA ....(((((((..((.(((....)))..))..))...((........(((((........)..((((...)))).))))...((((((((..(....)..))))))))....))))))). ( -37.30) >pfo.3002 5728541 120 - 6438405 CACUGUUUCGGCUAGGGGGUCAUCCCGACUUACCAAACCGAUGCAAACUCCGAAUACCUACAAGUGCCGAGCAUGGGAGACACACGGCGGGUGCUAACGUCCGUCGUGAAAAGGGAAACA ....(((((((..((.(((....)))..))..))...((........(((((........)..((((...)))).))))...((((((((..(....)..))))))))....))))))). ( -37.30) >pst.2031 3871633 120 - 6397126 CACUGUUUCGGCUAGGGGGUCAUCCCGACUUACCAAACCGAUGCAAACUCCGAAUACCUACAAGUGCCGAGCAUGGGAGACACACGGCGGGUGCUAACGUCCGUCGUGAAAAGGGAAACA ....(((((((..((.(((....)))..))..))...((........(((((........)..((((...)))).))))...((((((((..(....)..))))))))....))))))). ( -37.30) >consensus CACUGUUUCGGCUAGGGGGUCAUCCCGACUUACCAAACCGAUGCAAACUCCGAAUACCUACAAGUGCCGAGCAUGGGAGACACACGGCGGGUGCUAACGUCCGUCGUGAAAAGGGAAACA ....(((((((..((.(((....)))..))..))...((........(((((.....).....((((...)))).))))...((((((((..(....)..))))))))....))))))). (-37.07 = -37.07 + 0.00)
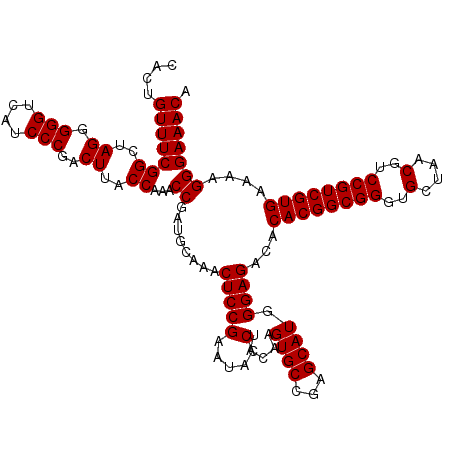
Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:04:14 2007