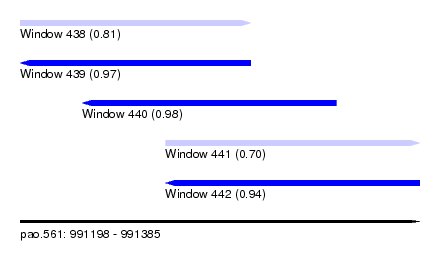
| Sequence ID | pao.561 |
|---|---|
| Location | 991,198 – 991,385 |
| Length | 187 |
| Max. P | 0.976826 |

| Location | 991,198 – 991,306 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.80 |
| Mean single sequence MFE | -38.73 |
| Consensus MFE | -28.93 |
| Energy contribution | -28.77 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.808216 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
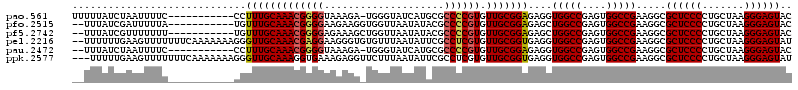
>pao.561 991198 108 + 6264404 UUUUUAUCUAAUUUUC-----------CCUUUGCAAACGGGGUAAAGA-UGGGUAUCAUGCGCCCCGUGUUGCGGAGAGGUGGCCGAGUGGCCGAAGGCGCUCCCCUGCUAAGGGAGUAC .............(((-----------((((.(((...((((....((-(....)))..((((((((.....))).....(((((....)))))..))))).))))))).)))))))... ( -40.80) >pfo.2515 1617232 107 - 6438405 --UUUAUCGAUUUUUA-----------UGUUUGCAAACGGGGAAGAAGGUGGUUAAUAUACGCCCCGUGUUGCGGAGAGCUGGCCGAGUGGCCGAAGGCGCUCCCCUGCUAAGGGAGUAC --..............-----------(.(((((((((((((......((.....)).....)))))).))))))).)(((((((....))))...)))((((((.......)))))).. ( -36.20) >pf5.2742 1860992 107 - 7074893 --UUUAUCGUUUUUUU-----------UGUUUGCAAACGGGGAGAAAGCUGGUUAAUAUACGCCCCGUGUUGCGGAGAGCUGGCCGAGUGGCCGAAGGCGCUCCCCUGCUAAGGGAGUAC --......((((((.(-----------(.(((((((((((((((....)).((......)).)))))).))))))).))..((((....))))))))))((((((.......)))))).. ( -36.30) >pel.2216 1731878 118 - 5888780 --UUUUUUGAAGUUUUUUUCAAAAAAAGGGUUGCAAACGAGGAAGGGUGUGUUUAAUAUUCGCCUCGUGUUGCGGUGAGGUGGCCGAGUGGCCGAAGGCGCUCCCCUGCUAAGGGAGUAU --((((((((((....))))))))))..(((..(..((((((..((((((.....)))))).))))))....((((......)))).)..)))......((((((.......)))))).. ( -41.40) >pau.2472 1877258 106 - 6537648 --UUUAUCUAAUUUUC-----------CCUUUGCAAACGGGGUAAAGA-UGGGUAUCAUGCGCCCCGUGUUGCGGAGAGGUGGCCGAGUGGCCGAAGGCGCUCCCCUGCUAAGGGAGUAC --...........(((-----------((((.(((...((((....((-(....)))..((((((((.....))).....(((((....)))))..))))).))))))).)))))))... ( -40.80) >ppk.2577 1101921 117 - 6181863 ---UUUUUGAAGUUUUUUUCAAAAAAAGGGUUGCAAAGGUGAAAGAGGUUCUUUAAUAUUCGCCUCGUGUUGCGGUGAGGUGGCCGAGUGGCCGAAGGCGCUCCCCUGCUAAGGGAGUAU ---(((((((((....)))))))))...(((..(...(((.((((.....))))......(((((((........))))))))))..)..)))......((((((.......)))))).. ( -36.90) >consensus __UUUAUCGAAUUUUU___________CGUUUGCAAACGGGGAAAAGG_UGGUUAAUAUACGCCCCGUGUUGCGGAGAGGUGGCCGAGUGGCCGAAGGCGCUCCCCUGCUAAGGGAGUAC .............................(((((((((((((....................)))))).)))))))....(((((....))))).....((((((.......)))))).. (-28.93 = -28.77 + -0.17)
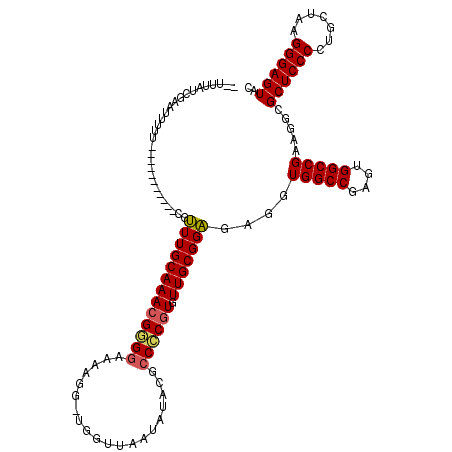
| Location | 991,198 – 991,306 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.80 |
| Mean single sequence MFE | -35.07 |
| Consensus MFE | -27.91 |
| Energy contribution | -27.88 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.33 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969548 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.561 991198 108 - 6264404 GUACUCCCUUAGCAGGGGAGCGCCUUCGGCCACUCGGCCACCUCUCCGCAACACGGGGCGCAUGAUACCCA-UCUUUACCCCGUUUGCAAAGG-----------GAAAAUUAGAUAAAAA ....((((((.(((((((.(((((...((((....))))......(((.....))))))))..(((....)-))....))))...))).))))-----------)).............. ( -40.90) >pfo.2515 1617232 107 + 6438405 GUACUCCCUUAGCAGGGGAGCGCCUUCGGCCACUCGGCCAGCUCUCCGCAACACGGGGCGUAUAUUAACCACCUUCUUCCCCGUUUGCAAACA-----------UAAAAAUCGAUAAA-- ..............(((((((......((((....)))).)))))))((((.((((((....................)))))))))).....-----------..............-- ( -32.25) >pf5.2742 1860992 107 + 7074893 GUACUCCCUUAGCAGGGGAGCGCCUUCGGCCACUCGGCCAGCUCUCCGCAACACGGGGCGUAUAUUAACCAGCUUUCUCCCCGUUUGCAAACA-----------AAAAAAACGAUAAA-- ..............(((((((......((((....)))).)))))))((((.((((((.(................).)))))))))).....-----------..............-- ( -33.49) >pel.2216 1731878 118 + 5888780 AUACUCCCUUAGCAGGGGAGCGCCUUCGGCCACUCGGCCACCUCACCGCAACACGAGGCGAAUAUUAAACACACCCUUCCUCGUUUGCAACCCUUUUUUUGAAAAAAACUUCAAAAAA-- ...((((((.....)))))).......((((....))))........((((.((((((....................))))))))))......(((((((((......)))))))))-- ( -32.85) >pau.2472 1877258 106 + 6537648 GUACUCCCUUAGCAGGGGAGCGCCUUCGGCCACUCGGCCACCUCUCCGCAACACGGGGCGCAUGAUACCCA-UCUUUACCCCGUUUGCAAAGG-----------GAAAAUUAGAUAAA-- ....((((((.(((((((.(((((...((((....))))......(((.....))))))))..(((....)-))....))))...))).))))-----------))............-- ( -40.90) >ppk.2577 1101921 117 + 6181863 AUACUCCCUUAGCAGGGGAGCGCCUUCGGCCACUCGGCCACCUCACCGCAACACGAGGCGAAUAUUAAAGAACCUCUUUCACCUUUGCAACCCUUUUUUUGAAAAAAACUUCAAAAA--- ...........(((((((..(((((..((((....)))).......((.....)))))))......((((.....))))..))))))).......((((((((......))))))))--- ( -30.00) >consensus GUACUCCCUUAGCAGGGGAGCGCCUUCGGCCACUCGGCCACCUCUCCGCAACACGGGGCGAAUAUUAACCA_CCUCUUCCCCGUUUGCAAACA___________AAAAAUUCGAUAAA__ ...((((((.....)))))).......((((....))))........((((.((((((....................))))))))))................................ (-27.91 = -27.88 + -0.03)
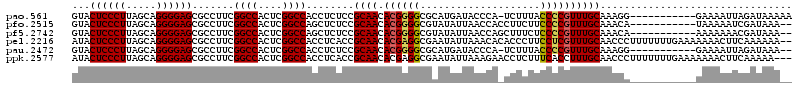
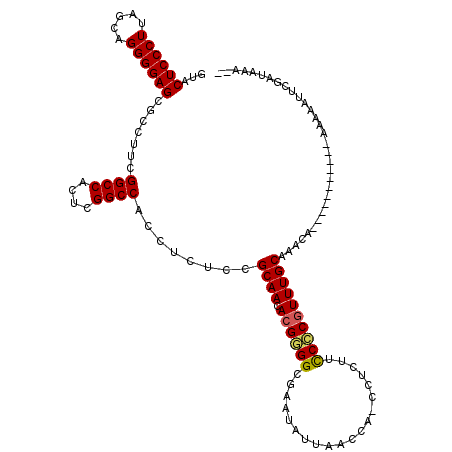
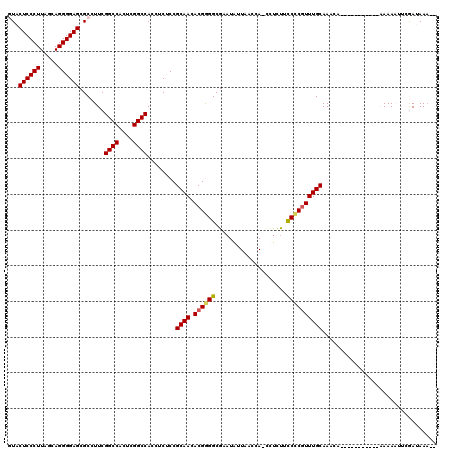
| Location | 991,227 – 991,346 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.72 |
| Mean single sequence MFE | -49.94 |
| Consensus MFE | -42.59 |
| Energy contribution | -42.03 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.976826 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.561 991227 119 - 6264404 GGAGAGGGGGGGAUUCGAACCCCCGACACCCUUUUGAGGUGUACUCCCUUAGCAGGGGAGCGCCUUCGGCCACUCGGCCACCUCUCCGCAACACGGGGCGCAUGAUACCCA-UCUUUACC ((((((((((((.......)))))...........((((.(..((((((.....))))))..)))))((((....)))).))))))).......(((..........))).-........ ( -53.00) >pfo.2515 1617259 120 + 6438405 GGAGAACGGGGGAUUCGAACCCCCGACACCCUUUUGAGGUGUACUCCCUUAGCAGGGGAGCGCCUUCGGCCACUCGGCCAGCUCUCCGCAACACGGGGCGUAUAUUAACCACCUUCUUCC ((((((.((((.......(((((((((((((....).))))).........((.(((((((......((((....)))).)))))))))....))))).)).......)).)))))))). ( -49.24) >pf5.2742 1861019 120 + 7074893 GGAGAACGGGGGAUUCGAACCCCCGACACCCUUUUGAGGUGUACUCCCUUAGCAGGGGAGCGCCUUCGGCCACUCGGCCAGCUCUCCGCAACACGGGGCGUAUAUUAACCAGCUUUCUCC ((((((((((((.......))))))((.((((...((((.(..((((((.....))))))..)))))((((....)))).((.....)).....)))).)).............)))))) ( -51.30) >pel.2216 1731916 120 + 5888780 GGUGAAGGGGGGAUUCGAACCCCCGAUACCCUUAUGAGGUAUACUCCCUUAGCAGGGGAGCGCCUUCGGCCACUCGGCCACCUCACCGCAACACGAGGCGAAUAUUAAACACACCCUUCC ((((..(((((........)))))((((......((((((...((((((.....)))))).......((((....)))))))))).(((........)))..)))).....))))..... ( -45.50) >pau.2472 1877285 119 + 6537648 GGAGAGGGGGGGAUUCGAACCCCCGACACCCUUUUGAGGUGUACUCCCUUAGCAGGGGAGCGCCUUCGGCCACUCGGCCACCUCUCCGCAACACGGGGCGCAUGAUACCCA-UCUUUACC ((((((((((((.......)))))...........((((.(..((((((.....))))))..)))))((((....)))).))))))).......(((..........))).-........ ( -53.00) >ppk.2577 1101958 120 + 6181863 GGUGAAGGGGGGAUUCGAACCCCCGAUACCCUUAUGAGGUAUACUCCCUUAGCAGGGGAGCGCCUUCGGCCACUCGGCCACCUCACCGCAACACGAGGCGAAUAUUAAAGAACCUCUUUC ((((..(((((........)))))..)))).....(((((...((((((.....))))))(((((..((((....)))).......((.....)))))))...........))))).... ( -47.60) >consensus GGAGAAGGGGGGAUUCGAACCCCCGACACCCUUUUGAGGUGUACUCCCUUAGCAGGGGAGCGCCUUCGGCCACUCGGCCACCUCUCCGCAACACGGGGCGAAUAUUAACCA_CCUCUUCC (((((.((((((.......)))))...........((((((..((((((.....)))))))))))).((((....)))).).)))))((........))..................... (-42.59 = -42.03 + -0.55)

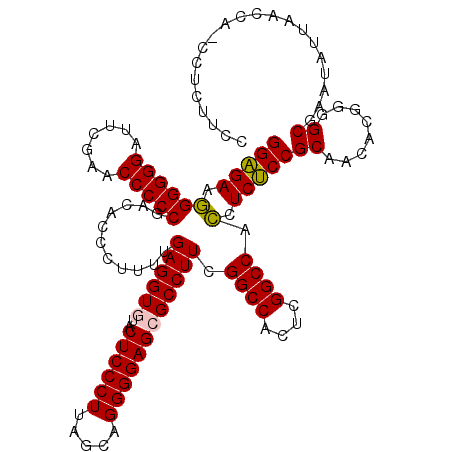
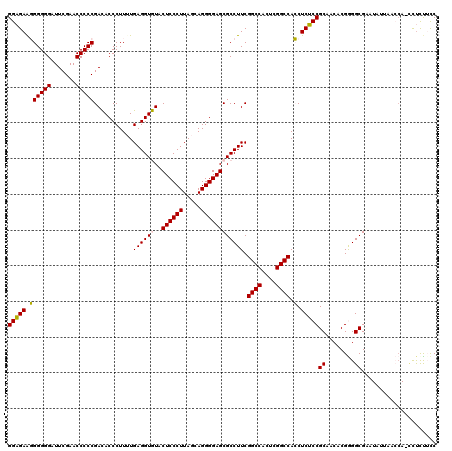
| Location | 991,266 – 991,385 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.42 |
| Mean single sequence MFE | -44.47 |
| Consensus MFE | -38.31 |
| Energy contribution | -37.87 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.699162 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.561 991266 119 + 6264404 UGGCCGAGUGGCCGAAGGCGCUCCCCUGCUAAGGGAGUACACCUCAAAAGGGUGUCGGGGGUUCGAAUCCCCCCCUCUCCGCCAUUCAAGUGUUAGAGGUUGAUUCAGGCGGCGA-AAGC ..((((((((((.(((((.((((((.......))))))((((((.....)))))).(((((.......)))))))).)).)))))))........(((.....))).))).((..-..)) ( -49.70) >pf5.2742 1861059 108 - 7074893 UGGCCGAGUGGCCGAAGGCGCUCCCCUGCUAAGGGAGUACACCUCAAAAGGGUGUCGGGGGUUCGAAUCCCCCGUUCUCCGCCAUU------------AUUUGCUUAGUACGUUGUAAUC .(((..((((((.((..((((((((.......))))))((((((.....)))))).(((((.......)))))))..)).))))))------------....)))............... ( -41.50) >pst.1101 2014816 107 + 6397126 UGGCCGAGUGGCCGAAGGCGCUCCCCUGCUAAGGGAGUACACCUCAAAAGGGUGUCGGGGGUUCGAAUCCCCCGUUCUCCGCCAUU------------AUUUAUGUAGCGCGUCA-GAUU ((((((((((((.((..((((((((.......))))))((((((.....)))))).(((((.......)))))))..)).))))))------------..........)).))))-.... ( -41.70) >pel.2216 1731956 108 - 5888780 UGGCCGAGUGGCCGAAGGCGCUCCCCUGCUAAGGGAGUAUACCUCAUAAGGGUAUCGGGGGUUCGAAUCCCCCCUUCACCGCCAUU------------AUUCGCUCAGGGCGCUUUAAGC .((((....)))).(((((((((.(((....)))((((((((((.....)))))).(((((........)))))............------------....)))).))))))))).... ( -42.50) >pau.2472 1877324 119 - 6537648 UGGCCGAGUGGCCGAAGGCGCUCCCCUGCUAAGGGAGUACACCUCAAAAGGGUGUCGGGGGUUCGAAUCCCCCCCUCUCCGCCAUUCAAGUGUUAGAGGUUGAUUCAGGCGGCGA-AAGC ..((((((((((.(((((.((((((.......))))))((((((.....)))))).(((((.......)))))))).)).)))))))........(((.....))).))).((..-..)) ( -49.70) >ppk.2577 1101998 108 - 6181863 UGGCCGAGUGGCCGAAGGCGCUCCCCUGCUAAGGGAGUAUACCUCAUAAGGGUAUCGGGGGUUCGAAUCCCCCCUUCACCGCCAUU------------AUUUGCGUAGGGCGCUGUAAAC .((((....))))(((((.((((((.......))))))((((((.....)))))).((((((....)))))))))))..((((.((------------((....))))))))........ ( -41.70) >consensus UGGCCGAGUGGCCGAAGGCGCUCCCCUGCUAAGGGAGUACACCUCAAAAGGGUGUCGGGGGUUCGAAUCCCCCCUUCUCCGCCAUU____________AUUUACUCAGGGCGCUA_AAGC .((((....))))...(((((((((.......))))))((((((.....)))))).(((((.......))))).......)))..................................... (-38.31 = -37.87 + -0.44)
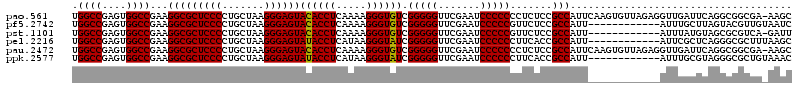
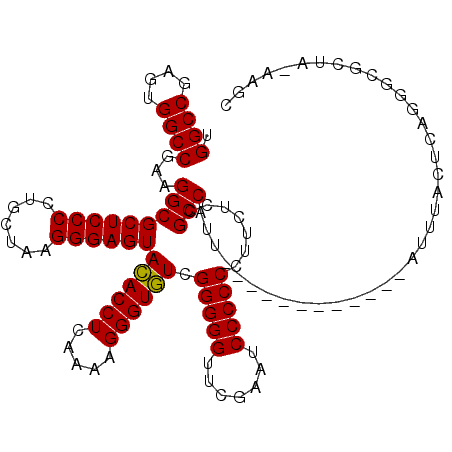
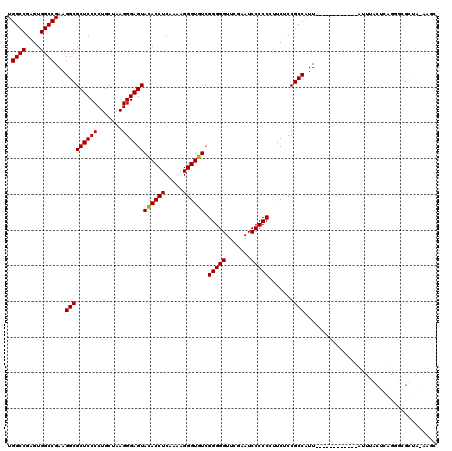
| Location | 991,266 – 991,385 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.42 |
| Mean single sequence MFE | -44.87 |
| Consensus MFE | -40.11 |
| Energy contribution | -39.67 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938767 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.561 991266 119 - 6264404 GCUU-UCGCCGCCUGAAUCAACCUCUAACACUUGAAUGGCGGAGAGGGGGGGAUUCGAACCCCCGACACCCUUUUGAGGUGUACUCCCUUAGCAGGGGAGCGCCUUCGGCCACUCGGCCA ...(-((.(((((....((((..........))))..))))).)))(((((........)))))...........((((.(..((((((.....))))))..)))))((((....)))). ( -48.10) >pf5.2742 1861059 108 + 7074893 GAUUACAACGUACUAAGCAAAU------------AAUGGCGGAGAACGGGGGAUUCGAACCCCCGACACCCUUUUGAGGUGUACUCCCUUAGCAGGGGAGCGCCUUCGGCCACUCGGCCA ......................------------...((((.....((((((.......))))))((((((....).))))).((((((.....))))))))))...((((....)))). ( -42.50) >pst.1101 2014816 107 - 6397126 AAUC-UGACGCGCUACAUAAAU------------AAUGGCGGAGAACGGGGGAUUCGAACCCCCGACACCCUUUUGAGGUGUACUCCCUUAGCAGGGGAGCGCCUUCGGCCACUCGGCCA ....-.................------------...((((.....((((((.......))))))((((((....).))))).((((((.....))))))))))...((((....)))). ( -42.50) >pel.2216 1731956 108 + 5888780 GCUUAAAGCGCCCUGAGCGAAU------------AAUGGCGGUGAAGGGGGGAUUCGAACCCCCGAUACCCUUAUGAGGUAUACUCCCUUAGCAGGGGAGCGCCUUCGGCCACUCGGCCA (((.....(((.....)))...------------...)))((((..(((((........))))).((((((....).))))).((((((.....))))))))))...((((....)))). ( -44.70) >pau.2472 1877324 119 + 6537648 GCUU-UCGCCGCCUGAAUCAACCUCUAACACUUGAAUGGCGGAGAGGGGGGGAUUCGAACCCCCGACACCCUUUUGAGGUGUACUCCCUUAGCAGGGGAGCGCCUUCGGCCACUCGGCCA ...(-((.(((((....((((..........))))..))))).)))(((((........)))))...........((((.(..((((((.....))))))..)))))((((....)))). ( -48.10) >ppk.2577 1101998 108 + 6181863 GUUUACAGCGCCCUACGCAAAU------------AAUGGCGGUGAAGGGGGGAUUCGAACCCCCGAUACCCUUAUGAGGUAUACUCCCUUAGCAGGGGAGCGCCUUCGGCCACUCGGCCA .......(((.....)))....------------...((((.....(((((........))))).((((((....).))))).((((((.....))))))))))...((((....)))). ( -43.30) >consensus GCUU_UAGCGCCCUAAACAAAU____________AAUGGCGGAGAAGGGGGGAUUCGAACCCCCGACACCCUUUUGAGGUGUACUCCCUUAGCAGGGGAGCGCCUUCGGCCACUCGGCCA .....................................((((......(((((.......))))).((((((....).))))).((((((.....))))))))))...((((....)))). (-40.11 = -39.67 + -0.44)

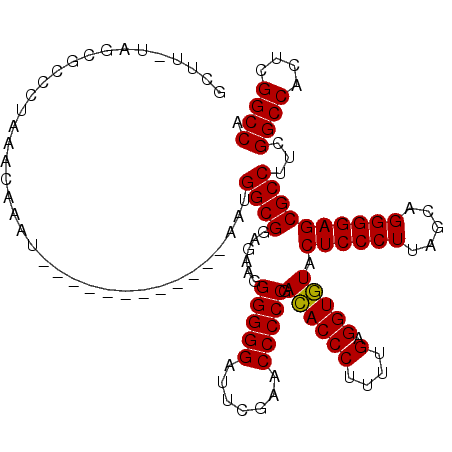
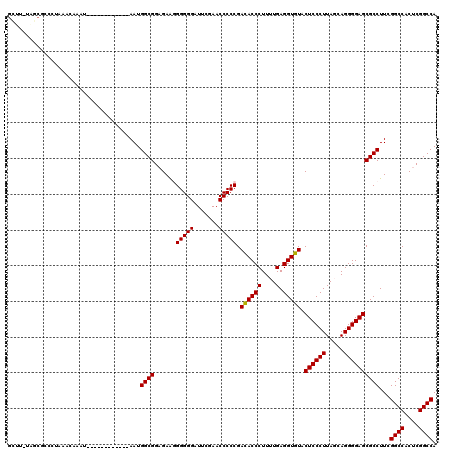
Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:11:21 2007