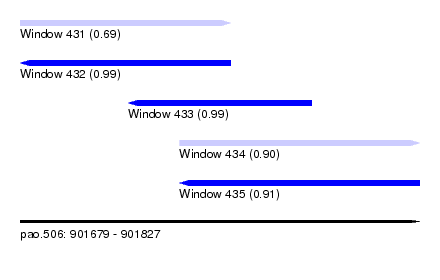
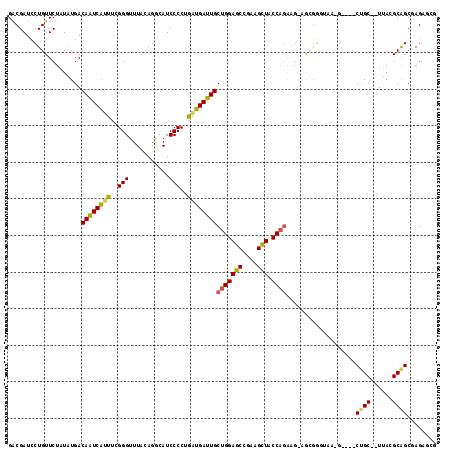
| Sequence ID | pao.506 |
|---|---|
| Location | 901,679 – 901,827 |
| Length | 148 |
| Max. P | 0.992278 |

| Location | 901,679 – 901,757 |
|---|---|
| Length | 78 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 75.09 |
| Mean single sequence MFE | -35.77 |
| Consensus MFE | -20.74 |
| Energy contribution | -20.28 |
| Covariance contribution | -0.45 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.691071 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
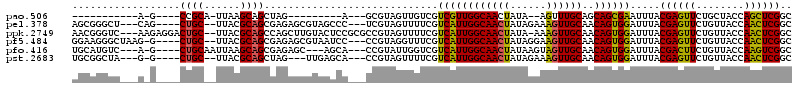
>pao.506 901679 78 + 6264404 GGCGAUCCUGUUCUAUCUGACAGUCGUUUCGGGUUUACAGGCAUCCCCUAGCGACUGCU-----------------------------A-G----CCGCA-UUAAGCAGCUAG---- ((.((((((((...((((((........))))))..))))).))).)).........((-----------------------------(-(----(.((.-....)).)))))---- ( -24.90) >pss.2544 1093647 110 - 6093698 GACGAUCCUGUUCUAUAUGACAAUCAUUUCGGGUUUACAGGCAUCCCCUGAUGAUUGCUGGAGCCGAAGCUACCAGAAG-GAAGGCUAGCA----CCGC--UUACGCGGCGAGAGCG .....((((...........((((((((..(((...........)))..))))))))(((((((....))).)))).))-))..(((..(.----((((--....)))).)..))). ( -37.40) >pel.378 791534 110 + 5888780 GACGAUCCUGUUCUGUAUGACAAUCAUUUCGGGUUUACAGGCAUCCCCUGAUGAUUGCUGGAGCCGAAGCUACCAGGAG-AGCGGGCUCAG----CUGC--UUACGCAGCGAGAGCG .......(((((((......((((((((..(((...........)))..))))))))(((((((....))).)))).))-)))))((((.(----((((--....)))))..)))). ( -44.60) >ppk.2749 791616 115 - 6181863 GACGAUCCUGUUCUGUAUGACAAUCAGUUCGGGUUUACAGGCAUCCCCUAGUGAUUGCUGGAGCCGAAGCUACCAGAAGCAACGGGUCAAGAGGACUGC--UUACGCAGCCAGCUUG ...((.((((..(((.((....)))))..)))).)).(((((...(((...((....(((((((....))).))))...))..)))......((.((((--....)))))).))))) ( -36.20) >pfo.416 878990 110 + 6438405 GGCGAUUCUGUUCUAUAUGACAAUCAUUUUGGGUUUACAGACAUCCCCUGAUGAUUGCUGGACCCGAAGGUACCAGGAG-UGCAUGUCA-G----CUGCAAUUAAGCAGCGAGAGC- .........(((((...(((((....((((((((((((((.((((....)))).))).)))))))))))((((.....)-))).)))))-(----((((......))))).)))))- ( -38.00) >pst.2683 1310716 106 - 6397126 GACGAUCCUGUUCUAUAUGACAAUCAUUUUGGGUUUACAGGCAUCCCCUGAUGAUUGCUGGAGCCGAAGCUACCAGAAG-UGCGGCUAG-G----CUGC--UUACGCAGCUAG---U .......((((.((......((((((((..(((...........)))..))))))))(((((((....))).)))).))-.))))...(-(----((((--....))))))..---. ( -33.50) >consensus GACGAUCCUGUUCUAUAUGACAAUCAUUUCGGGUUUACAGGCAUCCCCUGAUGAUUGCUGGAGCCGAAGCUACCAGAAG_AGCGGGUAA_G____CUGC__UUACGCAGCGAGAGCG ....................((((((((..(((...........)))..))))))))(((((((....))).))))...................((((......))))........ (-20.74 = -20.28 + -0.45)

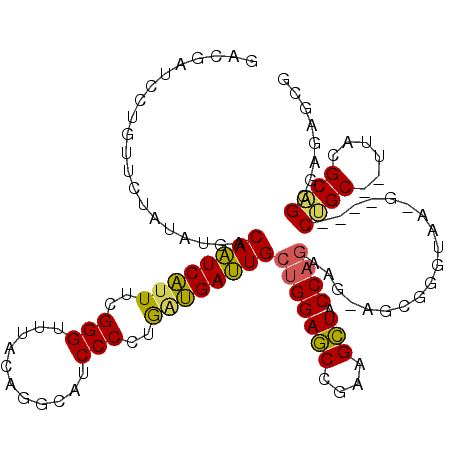
| Location | 901,679 – 901,757 |
|---|---|
| Length | 78 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 75.09 |
| Mean single sequence MFE | -37.31 |
| Consensus MFE | -23.44 |
| Energy contribution | -23.18 |
| Covariance contribution | -0.26 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.63 |
| SVM decision value | 2.32 |
| SVM RNA-class probability | 0.992278 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.506 901679 78 - 6264404 ----CUAGCUGCUUAA-UGCGG----C-U-----------------------------AGCAGUCGCUAGGGGAUGCCUGUAAACCCGAAACGACUGUCAGAUAGAACAGGAUCGCC ----((((((((....-.))))----)-)-----------------------------))((((((...(((..((....))..)))....))))))...(((........)))... ( -26.80) >pss.2544 1093647 110 + 6093698 CGCUCUCGCCGCGUAA--GCGG----UGCUAGCCUUC-CUUCUGGUAGCUUCGGCUCCAGCAAUCAUCAGGGGAUGCCUGUAAACCCGAAAUGAUUGUCAUAUAGAACAGGAUCGUC .(((..((((((....--))))----))..)))..((-((.((((.(((....)))))))(((((((..(((..((....))..)))...)))))))...........))))..... ( -43.30) >pel.378 791534 110 - 5888780 CGCUCUCGCUGCGUAA--GCAG----CUGAGCCCGCU-CUCCUGGUAGCUUCGGCUCCAGCAAUCAUCAGGGGAUGCCUGUAAACCCGAAAUGAUUGUCAUACAGAACAGGAUCGUC .((((..(((((....--))))----).))))..((.-.((((((.(((....))).).((((((((..(((..((....))..)))...)))))))).........)))))..)). ( -43.50) >ppk.2749 791616 115 + 6181863 CAAGCUGGCUGCGUAA--GCAGUCCUCUUGACCCGUUGCUUCUGGUAGCUUCGGCUCCAGCAAUCACUAGGGGAUGCCUGUAAACCCGAACUGAUUGUCAUACAGAACAGGAUCGUC ((((..((((((....--))))))..)))).(((..((.((((((.(((....))))))).)).))...)))(((.(((((........((.....))........))))))))... ( -40.69) >pfo.416 878990 110 - 6438405 -GCUCUCGCUGCUUAAUUGCAG----C-UGACAUGCA-CUCCUGGUACCUUCGGGUCCAGCAAUCAUCAGGGGAUGUCUGUAAACCCAAAAUGAUUGUCAUAUAGAACAGAAUCGCC -((....(((((......))))----)-((((.....-...((((.(((....))))))).((((((...(((...........)))...))))))))))..............)). ( -33.30) >pst.2683 1310716 106 + 6397126 A---CUAGCUGCGUAA--GCAG----C-CUAGCCGCA-CUUCUGGUAGCUUCGGCUCCAGCAAUCAUCAGGGGAUGCCUGUAAACCCAAAAUGAUUGUCAUAUAGAACAGGAUCGUC .---((((((((....--))))----.-))))((...-...((((.(((....)))))))(((((((..(((..((....))..)))...)))))))............))...... ( -36.30) >consensus CGCUCUCGCUGCGUAA__GCAG____C_UGACCCGCA_CUUCUGGUAGCUUCGGCUCCAGCAAUCAUCAGGGGAUGCCUGUAAACCCGAAAUGAUUGUCAUAUAGAACAGGAUCGUC ........((((......))))...................((((.(((....)))))))(((((((..(((..((....))..)))...))))))).................... (-23.44 = -23.18 + -0.26)

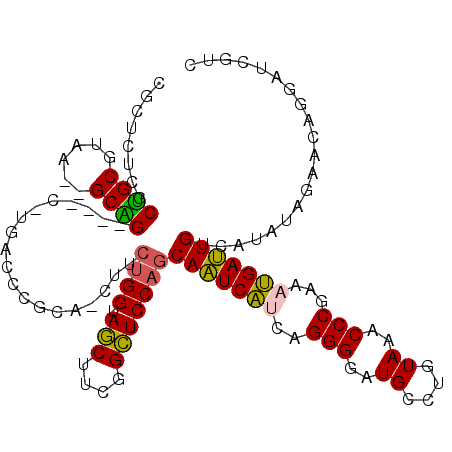
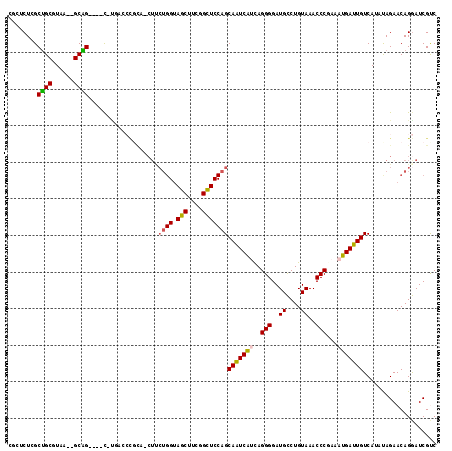
| Location | 901,719 – 901,787 |
|---|---|
| Length | 68 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 70.49 |
| Mean single sequence MFE | -35.75 |
| Consensus MFE | -16.43 |
| Energy contribution | -16.33 |
| Covariance contribution | -0.09 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.46 |
| SVM decision value | 2.16 |
| SVM RNA-class probability | 0.989431 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.506 901719 68 - 6264404 CU--UAUAGUUGCCAACGACGACAACUACGC---U---------CUAGCUGCUUAA-UGCGG----C-U-----------------------------AGCAGUCGCUAGGGGAUGC ..--....((((....))))..(..((((((---(---------((((((((....-.))))----)-)-----------------------------)).)).)).)))..).... ( -22.20) >pel.378 791574 107 - 5888780 UUUCUAUAGUUGCCAAUGACGAAAACUACGA---GGGCUACGCUCUCGCUGCGUAA--GCAG----CUGAGCCCGCU-CUCCUGGUAGCUUCGGCUCCAGCAAUCAUCAGGGGAUGC ............((..(((.((.......((---((((...((((..(((((....--))))----).))))..)))-)))((((.(((....)))))))...)).)))..)).... ( -44.20) >ppk.2749 791656 114 + 6181863 UUU-UAUAGUUGCCAAUGACGAAAACUACGGCGCGGAGUACAAGCUGGCUGCGUAA--GCAGUCCUCUUGACCCGUUGCUUCUGGUAGCUUCGGCUCCAGCAAUCACUAGGGGAUGC ...-....(((.((...............(((((((.((.((((..((((((....--))))))..))))))))).)))).((((.(((....))))))).........)).))).. ( -41.70) >pfo.416 879030 105 - 6438405 UACUUAUAGUUGCCAAUGACGACCAAUACGG---UGCU---GCUCUCGCUGCUUAAUUGCAG----C-UGACAUGCA-CUCCUGGUACCUUCGGGUCCAGCAAUCAUCAGGGGAUGU ............((..(((.((.......((---((((---(.((..(((((......))))----)-.)))).)))-)).((((.(((....)))))))...)).)))..)).... ( -34.60) >psp.2452 1124262 103 + 5928787 UUUCUAUAGUUGCCAAUGACGAAAACUACGG---UGCUCAA---CUAGCUGCGUAA--GCAG----C-CUAGCCGCA-CUUCUGGUAGCUUCGGCUCCAGCAAUCAUCAGGGGAUGU ............((..(((.((.......((---(((....---((((((((....--))))----.-))))..)))-)).((((.(((....)))))))...)).)))..)).... ( -35.90) >pst.2683 1310756 103 + 6397126 UUUCUAUAGUUGCCAAUGACGAAAACUACGG---UGCUCAA---CUAGCUGCGUAA--GCAG----C-CUAGCCGCA-CUUCUGGUAGCUUCGGCUCCAGCAAUCAUCAGGGGAUGC ............((..(((.((.......((---(((....---((((((((....--))))----.-))))..)))-)).((((.(((....)))))))...)).)))..)).... ( -35.90) >consensus UUUCUAUAGUUGCCAAUGACGAAAACUACGG___UGCU_A____CUAGCUGCGUAA__GCAG____C_UGACCCGCA_CUUCUGGUAGCUUCGGCUCCAGCAAUCAUCAGGGGAUGC ........(((.((..(((.............................((((......))))...................((((.(((....)))))))...)))...)).))).. (-16.43 = -16.33 + -0.09)
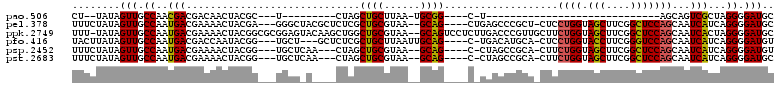
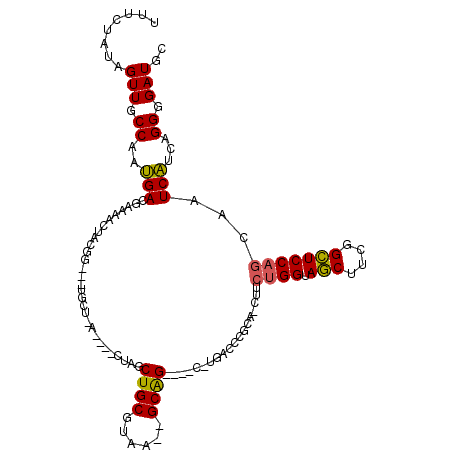
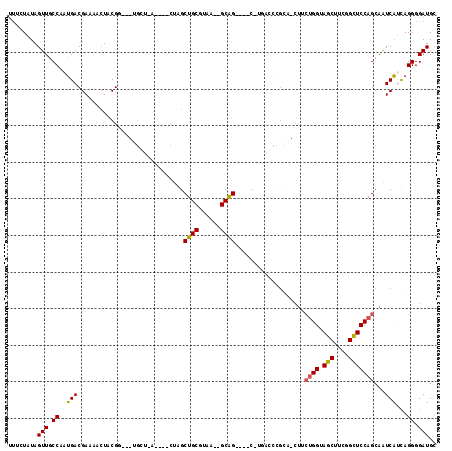
| Location | 901,738 – 901,827 |
|---|---|
| Length | 89 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.00 |
| Mean single sequence MFE | -35.13 |
| Consensus MFE | -20.28 |
| Energy contribution | -19.65 |
| Covariance contribution | -0.63 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.898194 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
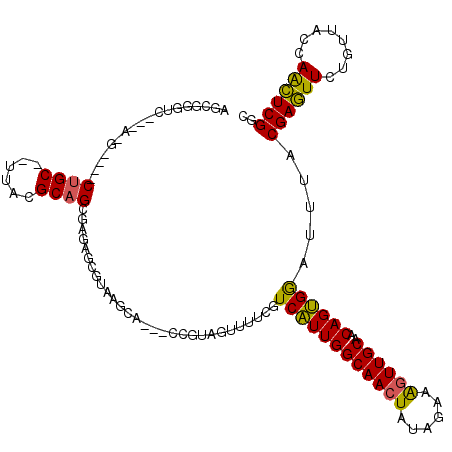
>pao.506 901738 89 + 6264404 -----------A-G----CCGCA-UUAAGCAGCUAG---------A---GCGUAGUUGUCGUCGUUGGCAACUAUA--AGUUUGCAGCAGCGAAUUUACGAGUUCUGCUACCAGCUCGGC -----------.-(----(((..-...((((((.((---------(---((((((((((((....))))))))))(--(((((((....))))))))....))))))))....))))))) ( -32.50) >pel.378 791613 108 + 5888780 AGCGGGCU---CAG----CUGC--UUACGCAGCGAGAGCGUAGCCC---UCGUAGUUUUCGUCAUUGGCAACUAUAGAAAGUUGCAACAGUGGAUUUACGAGUUCUGUUACCAACUCGGC .((.((((---(.(----((((--....)))))..))))(((((.(---((((((......((((((((((((......))))))..))))))..)))))))....))))).....).)) ( -39.40) >ppk.2749 791696 114 - 6181863 AACGGGUC---AAGAGGACUGC--UUACGCAGCCAGCUUGUACUCCGCGCCGUAGUUUUCGUCAUUGGCAACUAUA-AAAGUUGCAACAGUGGAUUUACGAGUUCUGUUACCAACUCGGC ..((((((---(((.((.((((--....))))))..)))).)).))).(((((((......((((((((((((...-..))))))..))))))..))))(((((........)))))))) ( -37.40) >pf5.484 938531 110 + 7074893 GGAAGGGCUAAG-G----CUGC--UUACGCAGCGAGAGCGUAAUCC---CCGUAGGUUUCGUCAUUGGCAACUAUAGGAAGUUGCAACAGUGGAUUUACGAGUUCUGUUACCAACUCGGC (((.(.(((...-(----((((--....)))))...))).)..)))---(((((((((....(((((((((((......))))))..))))))))))))(((((........))))))). ( -34.20) >pfo.416 879069 106 + 6438405 UGCAUGUC---A-G----CUGCAAUUAAGCAGCGAGAGC---AGCA---CCGUAUUGGUCGUCAUUGGCAACUAUAAGUAGUUGCAACAGUGGAUUUACGACUUCUGUUACCAAGUCGGC (((.(((.---.-(----((((......)))))....))---))))---(((..(((((........(((((((....)))))))(((((.((........)).))))))))))..))). ( -34.50) >pst.2683 1310795 104 - 6397126 UGCGGCUA---G-G----CUGC--UUACGCAGCUAG---UUGAGCA---CCGUAGUUUUCGUCAUUGGCAACUAUAGAAAGUUGCAACAGUGGAUUUACGAGUUCUGUUACCAACUCGGC ..((((((---(-.----((((--....))))))))---)))....---((((((......((((((((((((......))))))..))))))..))))(((((........))))))). ( -32.80) >consensus AGCGGGUC___A_G____CUGC__UUACGCAGCGAGAGCGUAAGCA___CCGUAGUUUUCGUCAUUGGCAACUAUAGAAAGUUGCAACAGUGGAUUUACGAGUUCUGUUACCAACUCGGC ..................((((......)))).............................((((((((((((......))))))..)))))).....((((((........)))))).. (-20.28 = -19.65 + -0.63)
| Location | 901,738 – 901,827 |
|---|---|
| Length | 89 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.00 |
| Mean single sequence MFE | -33.74 |
| Consensus MFE | -15.74 |
| Energy contribution | -15.55 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.47 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.912338 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
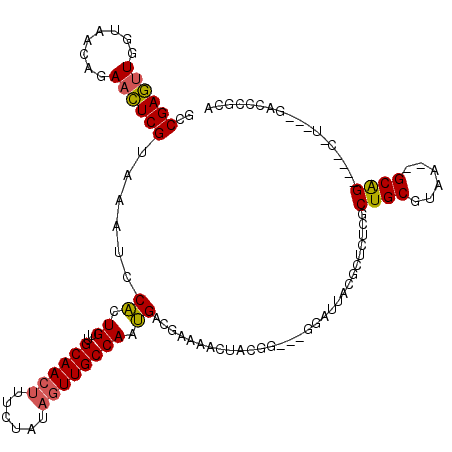
>pao.506 901738 89 - 6264404 GCCGAGCUGGUAGCAGAACUCGUAAAUUCGCUGCUGCAAACU--UAUAGUUGCCAACGACGACAACUACGC---U---------CUAGCUGCUUAA-UGCGG----C-U----------- ...((((..(((((((....((......))))))))).....--..((((((.(......).)))))).))---)---------).((((((....-.))))----)-)----------- ( -29.20) >pel.378 791613 108 - 5888780 GCCGAGUUGGUAACAGAACUCGUAAAUCCACUGUUGCAACUUUCUAUAGUUGCCAAUGACGAAAACUACGA---GGGCUACGCUCUCGCUGCGUAA--GCAG----CUG---AGCCCGCU (((...(((....)))..((((((.......((((((((((......))))))....)))).....)))))---))))...((((..(((((....--))))----).)---)))..... ( -35.90) >ppk.2749 791696 114 + 6181863 GCCGAGUUGGUAACAGAACUCGUAAAUCCACUGUUGCAACUUU-UAUAGUUGCCAAUGACGAAAACUACGGCGCGGAGUACAAGCUGGCUGCGUAA--GCAGUCCUCUU---GACCCGUU ((((((((.(((((((..............)))))((((((..-...)))))).....))...)))).))))((((.((.((((..((((((....--))))))..)))---))))))). ( -41.64) >pf5.484 938531 110 - 7074893 GCCGAGUUGGUAACAGAACUCGUAAAUCCACUGUUGCAACUUCCUAUAGUUGCCAAUGACGAAACCUACGG---GGAUUACGCUCUCGCUGCGUAA--GCAG----C-CUUAGCCCUUCC ..((((((........))))))..(((((.((((.((((((......))))))..............))))---)))))..(((...(((((....--))))----)-...)))...... ( -33.06) >pfo.416 879069 106 - 6438405 GCCGACUUGGUAACAGAAGUCGUAAAUCCACUGUUGCAACUACUUAUAGUUGCCAAUGACGACCAAUACGG---UGCU---GCUCUCGCUGCUUAAUUGCAG----C-U---GACAUGCA ((((..((((((((((..(........)..)))))(((((((....)))))))........)))))..)))---)(((---(.((..(((((......))))----)-.---)))).)). ( -32.30) >pst.2683 1310795 104 + 6397126 GCCGAGUUGGUAACAGAACUCGUAAAUCCACUGUUGCAACUUUCUAUAGUUGCCAAUGACGAAAACUACGG---UGCUCAA---CUAGCUGCGUAA--GCAG----C-C---UAGCCGCA ((((((((.(((((((..............)))))((((((......)))))).....))...)))).)))---)((....---((((((((....--))))----.-)---)))..)). ( -30.34) >consensus GCCGAGUUGGUAACAGAACUCGUAAAUCCACUGUUGCAACUUUCUAUAGUUGCCAAUGACGAAAACUACGG___GGAUUACGCUCUCGCUGCGUAA__GCAG____C_U___GACCCGCA ..((((((........))))))......((.((..((((((......)))))))).))..............................((((......)))).................. (-15.74 = -15.55 + -0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:11:14 2007