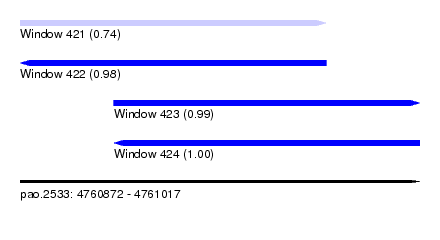
| Sequence ID | pao.2533 |
|---|---|
| Location | 4,760,872 – 4,761,017 |
| Length | 145 |
| Max. P | 0.995318 |

| Location | 4,760,872 – 4,760,983 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.90 |
| Mean single sequence MFE | -33.22 |
| Consensus MFE | -19.56 |
| Energy contribution | -20.28 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.743410 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2533 4760872 111 + 6264404 GGGCUUAACGCUCCUGAUACAA-AAG-AAUAAGCCUU-CAACUUCAAUU----CGCCCAUAAA-CGCCAGGCGAA-ACAAACCCAGGCUCAGGGGAGCCGGGUAUUCUAGAGAUCACGCC ((((.....)))).((((....-.((-(((.......-.........((----((((......-.....))))))-....((((.(((((....))))))))))))))....)))).... ( -30.70) >pfo.2990 5698839 113 + 6438405 --GCUAGACGCUCCUAAUACAG-AAA-AAUUAGCCUUGCGGCUACUACUU-GUCGCCGAGAAA-UUCCGGGCAUA-AAAAACACGGGCUCAGGCGAGCCGGUCAUUCUAGACAUACCCCA --.(((((.(((((((((....-...-.))))).((((((((........-)))).))))...-....))))...-......((.(((((....))))).))...))))).......... ( -30.50) >pf5.3366 6355761 111 + 7074893 --GCUAGAUGCUCCUGAUACAA-AAAGAAUUAGCCUUGCGGCUACAACU--GUCGCCGAGAAA-UUCCGGGCA---AAAAACACGGGCUCAGGCGAGCCGGUCAUUCUAGACACACCCCA --.(((((((((((((((.(..-...).))))).((((((((.......--)))).))))...-....)))))---......((.(((((....))))).))...))))).......... ( -31.00) >ppk.270 5624402 110 - 6181863 --GCUAGAUGCUCCUGAUACAAGAAU-UAUUAGCCUUGCGGCUAUGACA--ACCACC-CGAAC-CGUUUGGGC---AAAAACCCAGGCUCAGGUGAGCCGGUCAUUCUAGACACAAGGCA --((.....))...............-.....(((((((((((......--..((((-.((..-.(((((((.---.....))))))))).)))))))))(((......))).)))))). ( -34.80) >pel.250 5365965 111 - 5888780 --GCUAGAUGCUCCUGAUACAAGAAU-UAUUAGCCUUGCGGCUAUCACA--ACCACC-CGAGCGCGCAGGGCA---AAAAACCCAGGCUCGGGUGAGCCGGUCAUUCUAGACACAAGGCA --((.....))...............-.....(((((((((((......--..((((-(((((.(...(((..---.....))).)))))))))))))))(((......))).)))))). ( -41.80) >pss.2750 5396686 115 + 6093698 --GCUAGACGCUCCUAAAACAA-AAU-AAUUAGCCUUGCGGCUACAGCUUGAUCGCCGAGAAC-UUCCGGGCAAGUAAAAACACGGGCUCAGGCGAGCCGGCAAUUCUAGUCCUACGCCG --((((((..............-...-...(((((....)))))..(((.(.(((((..(...-...)((((..((......))..)))).))))).).)))...))))))......... ( -30.50) >consensus __GCUAGACGCUCCUGAUACAA_AAU_AAUUAGCCUUGCGGCUACAACU__AUCGCCGAGAAA_CGCCGGGCA___AAAAACACAGGCUCAGGCGAGCCGGUCAUUCUAGACACAACCCA ...(((((....((................(((((....)))))..........(((............))).............(((((....)))))))....))))).......... (-19.56 = -20.28 + 0.72)


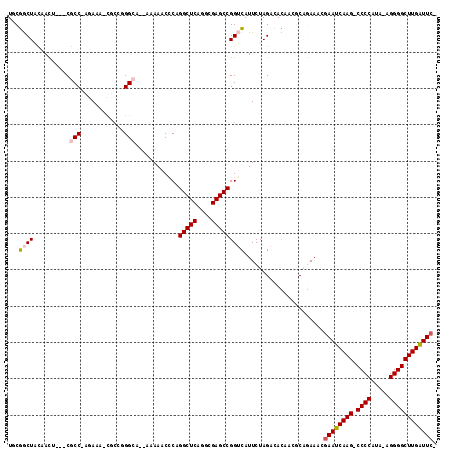
| Location | 4,760,872 – 4,760,983 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.90 |
| Mean single sequence MFE | -41.17 |
| Consensus MFE | -26.35 |
| Energy contribution | -26.50 |
| Covariance contribution | 0.15 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977252 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2533 4760872 111 - 6264404 GGCGUGAUCUCUAGAAUACCCGGCUCCCCUGAGCCUGGGUUUGU-UUCGCCUGGCG-UUUAUGGGCG----AAUUGAAGUUG-AAGGCUUAUU-CUU-UUGUAUCAGGAGCGUUAAGCCC (((.((((((((.((..(((((((((....))))).))))....-((((((((...-....))))))----))...((((..-...))))...-...-.....)).)))).)))).))). ( -35.50) >pfo.2990 5698839 113 - 6438405 UGGGGUAUGUCUAGAAUGACCGGCUCGCCUGAGCCCGUGUUUUU-UAUGCCCGGAA-UUUCUCGGCGAC-AAGUAGUAGCCGCAAGGCUAAUU-UUU-CUGUAUUAGGAGCGUCUAGC-- ..(((((((...(((((.((.(((((....))))).))))))).-)))))))....-......((((..-......(((((....)))))...-.((-((......))))))))....-- ( -36.60) >pf5.3366 6355761 111 - 7074893 UGGGGUGUGUCUAGAAUGACCGGCUCGCCUGAGCCCGUGUUUUU---UGCCCGGAA-UUUCUCGGCGAC--AGUUGUAGCCGCAAGGCUAAUUCUUU-UUGUAUCAGGAGCAUCUAGC-- ..(((((((((.(((((.((.(((((....))))).))))))).---.(((.((..-...)).))))))--((...(((((....)))))...))..-...........))))))...-- ( -35.70) >ppk.270 5624402 110 + 6181863 UGCCUUGUGUCUAGAAUGACCGGCUCACCUGAGCCUGGGUUUUU---GCCCAAACG-GUUCG-GGUGGU--UGUCAUAGCCGCAAGGCUAAUA-AUUCUUGUAUCAGGAGCAUCUAGC-- (((((((...(.((((((((.(((.(((((((((((((((....---)))))...)-)))))-))))))--)))).(((((....)))))...-))))).)...)))).)))......-- ( -49.90) >pel.250 5365965 111 + 5888780 UGCCUUGUGUCUAGAAUGACCGGCUCACCCGAGCCUGGGUUUUU---UGCCCUGCGCGCUCG-GGUGGU--UGUGAUAGCCGCAAGGCUAAUA-AUUCUUGUAUCAGGAGCAUCUAGC-- (((((((...(.(((((..(((((.((((((((((.((((....---.))))...).)))))-))))))--)).).(((((....)))))...-))))).)...)))).)))......-- ( -48.90) >pss.2750 5396686 115 - 6093698 CGGCGUAGGACUAGAAUUGCCGGCUCGCCUGAGCCCGUGUUUUUACUUGCCCGGAA-GUUCUCGGCGAUCAAGCUGUAGCCGCAAGGCUAAUU-AUU-UUGUUUUAGGAGCGUCUAGC-- (((.(((((..(((((..((.(((((....))))).))..)))))))))))))((.-(((((((((......))))(((((....)))))...-...-........))))).))....-- ( -40.40) >consensus UGCCGUAUGUCUAGAAUGACCGGCUCGCCUGAGCCCGGGUUUUU___UGCCCGGAA_GUUCUCGGCGAU__AGUUGUAGCCGCAAGGCUAAUU_AUU_UUGUAUCAGGAGCAUCUAGC__ ..........(((((...((.(((((....))))).))((((((...((((.((......)).)))).........(((((....)))))...............)))))).)))))... (-26.35 = -26.50 + 0.15)
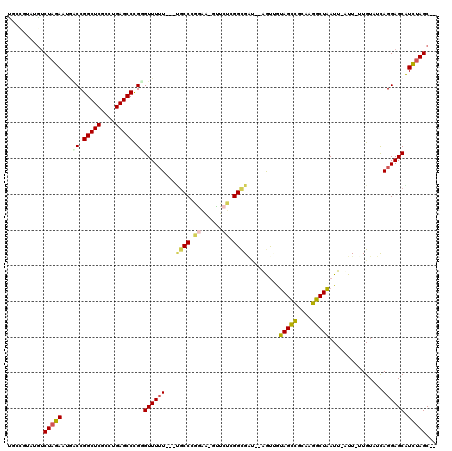
| Location | 4,760,906 – 4,761,017 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 75.61 |
| Mean single sequence MFE | -47.10 |
| Consensus MFE | -29.20 |
| Energy contribution | -29.15 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.08 |
| Mean z-score | -4.86 |
| Structure conservation index | 0.62 |
| SVM decision value | 2.36 |
| SVM RNA-class probability | 0.992898 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2533 4760906 111 + 6264404 U-CAACUUCAAUU---CGCCCAUAAA-CGCCAGGCGAAACAAACCCAGGCUCAGGGGAGCCGGGUAUUCUAGAGAUCACGCCAAAACGAAUCAAG-CCCCAUG-AGGGGCUUGAUUCG .-....(((..((---((((......-.....))))))....((((.(((((....)))))))))......)))............(((((((((-((((...-.))))))))))))) ( -47.00) >pfo.2990 5698871 114 + 6438405 UGCGGCUACUACUUGUCGCCGAGAAA-UUCCGGGCAUAAAAAACACGGGCUCAGGCGAGCCGGUCAUUCUAGACAUACCCCAGAAAUGAAUCAAG-CCCCAAA-AGGGGCUUGAUUC- ..((((.((.....)).)))).....-(((.(((.............(((((....))))).(((......)))....))).)))..((((((((-((((...-.))))))))))))- ( -46.40) >pf5.3366 6355794 112 + 7074893 UGCGGCUACAACU-GUCGCCGAGAAA-UUCCGGGCA--AAAAACACGGGCUCAGGCGAGCCGGUCAUUCUAGACACACCCCAGAAAUGAAUCAAGCCCCCAAA-AGGGGCUUGAUUC- ..((((.((....-)).)))).....-(((.(((..--.........(((((....))))).(((......)))....))).)))..((((((((((((....-.))))))))))))- ( -45.70) >ppk.270 5624435 111 - 6181863 UGCGGCUAUGACA-ACCACC-CGAAC-CGUUUGGGC--AAAAACCCAGGCUCAGGUGAGCCGGUCAUUCUAGACACAAGGCAGAAACGAAACAAG-CCCCAUAUAGGGGCUUGUUUG- (((((((......-..((((-.((..-.(((((((.--.....))))))))).)))))))).(((......))).....)))......(((((((-((((.....))))))))))).- ( -43.70) >pel.250 5365998 112 - 5888780 UGCGGCUAUCACA-ACCACC-CGAGCGCGCAGGGCA--AAAAACCCAGGCUCGGGUGAGCCGGUCAUUCUAGACACAAGGCAGAAACGAAACAAG-CCCCAUAUAGGGGCUUGUUUC- (((((((......-..((((-(((((.(...(((..--.....))).)))))))))))))).(((......))).....))).....((((((((-((((.....))))))))))))- ( -52.80) >pau.426 5771736 111 - 6537648 U-CAACUUCAAUU---CGCCCAUAAA-CGCCAGGCGAAACAAACCCAGGCUCAGGGGAGCCGGGUAUUCUAGAGAUCGCGCCAAAACGAAUCAAG-CCCCAUG-AGGGGCUUGAUUCG .-....(((..((---((((......-.....))))))....((((.(((((....)))))))))......)))............(((((((((-((((...-.))))))))))))) ( -47.00) >consensus UGCGGCUACAACU___CGCC_AGAAA_CGCCGGGCA__AAAAACCCAGGCUCAGGCGAGCCGGUCAUUCUAGACACAACGCAGAAACGAAUCAAG_CCCCAUA_AGGGGCUUGAUUC_ ...((((..........(((............)))............(((((....)))))))))......................((((((((.((((.....)))))))))))). (-29.20 = -29.15 + -0.05)

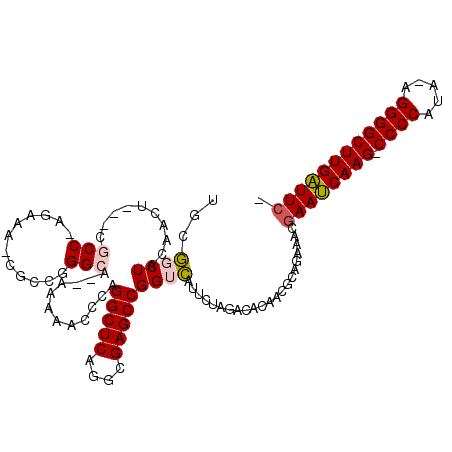
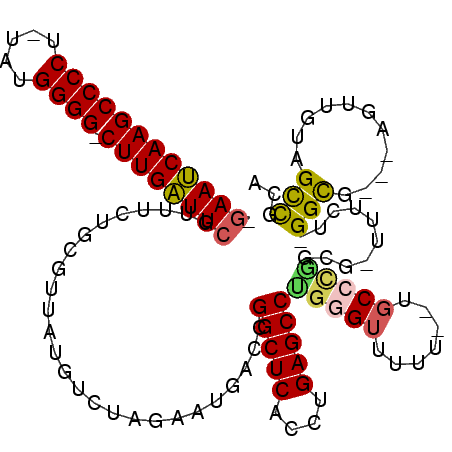
| Location | 4,760,906 – 4,761,017 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 75.61 |
| Mean single sequence MFE | -52.07 |
| Consensus MFE | -30.55 |
| Energy contribution | -30.00 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.28 |
| Mean z-score | -4.83 |
| Structure conservation index | 0.59 |
| SVM decision value | 2.56 |
| SVM RNA-class probability | 0.995318 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2533 4760906 111 - 6264404 CGAAUCAAGCCCCU-CAUGGGG-CUUGAUUCGUUUUGGCGUGAUCUCUAGAAUACCCGGCUCCCCUGAGCCUGGGUUUGUUUCGCCUGGCG-UUUAUGGGCG---AAUUGAAGUUG-A .((((((((((((.-...))))-))))))))(((((((.(....).)))))))(((((((((....))))).))))....((((((((...-....))))))---)).........-. ( -50.40) >pfo.2990 5698871 114 - 6438405 -GAAUCAAGCCCCU-UUUGGGG-CUUGAUUCAUUUCUGGGGUAUGUCUAGAAUGACCGGCUCGCCUGAGCCCGUGUUUUUUAUGCCCGGAA-UUUCUCGGCGACAAGUAGUAGCCGCA -((((((((((((.-...))))-))))))))..(((((((.((((...(((((.((.(((((....))))).))))))).)))))))))))-.....((((.((.....)).)))).. ( -52.60) >pf5.3366 6355794 112 - 7074893 -GAAUCAAGCCCCU-UUUGGGGGCUUGAUUCAUUUCUGGGGUGUGUCUAGAAUGACCGGCUCGCCUGAGCCCGUGUUUUU--UGCCCGGAA-UUUCUCGGCGAC-AGUUGUAGCCGCA -((((((((((((.-....))))))))))))..(((((((..(.(((......))))(((((....))))).........--..)))))))-.....((((.((-....)).)))).. ( -50.80) >ppk.270 5624435 111 + 6181863 -CAAACAAGCCCCUAUAUGGGG-CUUGUUUCGUUUCUGCCUUGUGUCUAGAAUGACCGGCUCACCUGAGCCUGGGUUUUU--GCCCAAACG-GUUCG-GGUGGU-UGUCAUAGCCGCA -.(((((((((((.....))))-)))))))...........((((.(((...((((.(((.(((((((((((((((....--)))))...)-)))))-))))))-)))))))).)))) ( -52.60) >pel.250 5365998 112 + 5888780 -GAAACAAGCCCCUAUAUGGGG-CUUGUUUCGUUUCUGCCUUGUGUCUAGAAUGACCGGCUCACCCGAGCCUGGGUUUUU--UGCCCUGCGCGCUCG-GGUGGU-UGUGAUAGCCGCA -((((((((((((.....))))-))))))))......((...(.(((......))))((((((((((((((.((((....--.))))...).)))))-))))..-......)))))). ( -55.50) >pau.426 5771736 111 + 6537648 CGAAUCAAGCCCCU-CAUGGGG-CUUGAUUCGUUUUGGCGCGAUCUCUAGAAUACCCGGCUCCCCUGAGCCUGGGUUUGUUUCGCCUGGCG-UUUAUGGGCG---AAUUGAAGUUG-A (((((((((((((.-...))))-)))))))))........(((..((......(((((((((....))))).))))....((((((((...-....))))))---))..))..)))-. ( -50.50) >consensus _GAAUCAAGCCCCU_UAUGGGG_CUUGAUUCGUUUCUGCGUUAUGUCUAGAAUGACCGGCUCACCUGAGCCUGGGUUUUU__UGCCCGGCG_UUUCU_GGCG___AGUUGUAGCCGCA .((((((((((((.....)))).))))))))..........................(((((....)))))(((((.......)))))..........(((...........)))... (-30.55 = -30.00 + -0.55)

Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:11:02 2007