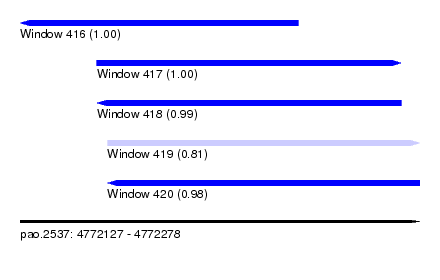
| Sequence ID | pao.2537 |
|---|---|
| Location | 4,772,127 – 4,772,278 |
| Length | 151 |
| Max. P | 0.996499 |

| Location | 4,772,127 – 4,772,232 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.33 |
| Mean single sequence MFE | -38.40 |
| Consensus MFE | -25.00 |
| Energy contribution | -25.17 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.54 |
| Mean z-score | -3.04 |
| Structure conservation index | 0.65 |
| SVM decision value | 2.58 |
| SVM RNA-class probability | 0.995456 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2537 4772127 105 - 6264404 UGACGGUGUACAAGAACCUCUUUACUAUUUUGUACCCCGAAAAUUGGCAGGGCGCACGCUCUGUCAUUU-CGUUUGU--------------GUGUCGUAAGACAAUCAGUGGAGCUAUAG ...(((.((((((((............)))))))).))).....(((((((((....))))))))).((-(..(((.--------------.((((....))))..)))..)))...... ( -32.80) >psp.2676 5231931 114 - 5928787 UGACUGGGGACGGGAUCCUCUUUAGACUCUUGUACCCCUAAAUUUGGCGGGGCUUGUGCCCUGUCAUUU--GCUUUUCUUGUAAGACAAUAGCGUCGCAAGACAA-CAGUGGAGCUA--- .....((((((((((..((....))..)))))).))))......(((((((((....)))))))))((.--.((..((((((..(((......)))))))))...-.))..))....--- ( -40.50) >pfo.2994 5710119 115 - 6438405 UGACUGGGGACAAGAUCCUCUUUAGACUCUUGAUCCCCUAAAUUUGGCGGGAAUUCGUUCCUGCCAUUU-UGCUUUUCUUGCAAGACAAUAGCGUCGCAAGACAA-CAGUGGAGCUA--- .....((((((((((..((....))..))))).)))))......(((((((((....)))))))))(((-..((..((((((..(((......)))))))))...-.))..)))...--- ( -41.20) >pf5.3370 6367070 99 - 7074893 UGACUGGGGGCAAGAUCCUCUUUAGACUCUUGUACCCCUAAAUUUGGUGGAGCUCCUGCUCUGCCAUUU-UGUUUAU----------------GUCGAAAGACAA-CAGUGGAGCUA--- (.(((((((((((((..((....))..)))))..))))(((((.(((..((((....))))..)))...-.)))))(----------------(((....)))).-)))).).....--- ( -37.10) >ppk.266 5635698 117 + 6181863 UGACUGGGUGCAAGAUCCUCUUUAGACUUUUGUACCCUUAAAUUUGGUGAGGCUCCGUCUCGCCAAUUUUUGGCUUUCUUGCAAGACAAUAGAGUCGCAAGACAAUCAGUGGAGCUA--- .....((((((((((..((....))..))))))))))..(((.((((((((((...)))))))))).)))((((((((((((..(((......))))))))).........))))))--- ( -39.40) >pel.246 5377262 117 + 5888780 UGACUGGGUGCAAGAUCCUCUUUAGACUUUUGUACCCUUAAAUUUGGUGAGGCUCCGUCUCGCCAAUUUUUGGCUUUCUUGCAAGACAAUAGAGUCGCAAGACAAUCAGUGGAGCUA--- .....((((((((((..((....))..))))))))))..(((.((((((((((...)))))))))).)))((((((((((((..(((......))))))))).........))))))--- ( -39.40) >consensus UGACUGGGGACAAGAUCCUCUUUAGACUCUUGUACCCCUAAAUUUGGCGGGGCUCCGGCUCUGCCAUUU_UGCUUUUCUUGCAAGACAAUAGAGUCGCAAGACAA_CAGUGGAGCUA___ .....((((((((((((.......)).)))))))))).......(((((((((....)))))))))...........................(((....)))................. (-25.00 = -25.17 + 0.17)

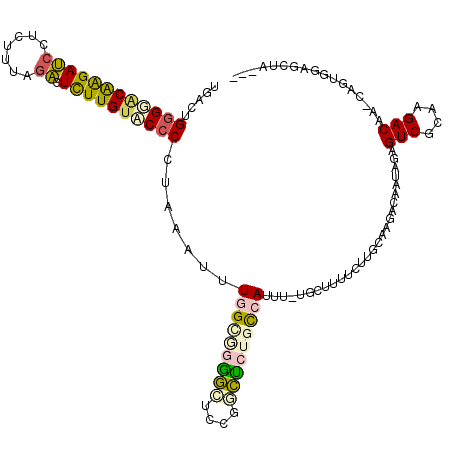
| Location | 4,772,156 – 4,772,271 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 68.78 |
| Mean single sequence MFE | -36.63 |
| Consensus MFE | -9.82 |
| Energy contribution | -9.52 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.62 |
| Mean z-score | -3.45 |
| Structure conservation index | 0.27 |
| SVM decision value | 2.71 |
| SVM RNA-class probability | 0.996499 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2537 4772156 115 + 6264404 ---ACAAACG-AAAUGACAGAGCGUGCGCCCUGCCAAUUUUCGGGGUACAAAAUAGUAAAGAGGUUCUUGUACACCGUCAAGACAAAGCCCCGGCUA-UCGGGAUAGCCGGGGCUUGUCG ---.....((-(((((.(((.((....)).))).))..)))))(((((((((((.........))).)))))).)).....(((((.((((((((((-(....)))))))))))))))). ( -47.10) >pau.422 5782985 115 - 6537648 ---ACAAACG-AAAUGACAGAGCGUGCGCCCUGCCAAUUUUCGGGGUACAAAAUAGUAAAGAGGUUCUUGUACACCGUCAAGACAAAGCCCCGGCUA-UCGGGAUAGCCGGGGCUUGUCG ---.....((-(((((.(((.((....)).))).))..)))))(((((((((((.........))).)))))).)).....(((((.((((((((((-(....)))))))))))))))). ( -47.10) >pfo.2994 5710155 97 + 6438405 AAGAAAAGCA-AAAUGGCAGGAACGAAUUCCCGCCAAAUUUAGGGGAUCAAGAGUCUAAAGAGGAUCUUGUCCCCAGUCAAGGCAAGGCCCCGAC-C-UC-------------------- ..........-...((((.((((....)))).))))......(((((.(((((..((....))..)))))))))).(((..(((...)))..)))-.-..-------------------- ( -32.50) >pf5.3370 6367093 94 + 7074893 ---AUAAACA-AAAUGGCAGAGCAGGAGCUCCACCAAAUUUAGGGGUACAAGAGUCUAAAGAGGAUCUUGCCCCCAGUCAAGGCAAGGCCCCGGC-C-UC-------------------- ---.......-...(((..((((....))))..)))......((((..(((((..((....))..))))).)))).((....)).((((....))-)-).-------------------- ( -29.30) >ppk.266 5635735 99 - 6181863 AAGAAAGCCAAAAAUUGGCGAGACGGAGCCUCACCAAAUUUAAGGGUACAAAAGUCUAAAGAGGAUCUUGCACCCAGUCAAGACGAGGCCCCGGC-CCUC-------------------- ......((((.....))))(((.(((.(((((...........((((.(((..((((.....)))).))).)))).((....))))))).)))..-.)))-------------------- ( -31.90) >pel.246 5377299 99 - 5888780 AAGAAAGCCAAAAAUUGGCGAGACGGAGCCUCACCAAAUUUAAGGGUACAAAAGUCUAAAGAGGAUCUUGCACCCAGUCAAGACGAGGCCCCGGC-CCUC-------------------- ......((((.....))))(((.(((.(((((...........((((.(((..((((.....)))).))).)))).((....))))))).)))..-.)))-------------------- ( -31.90) >consensus ___AAAAACA_AAAUGGCAGAGACGGAGCCCCACCAAAUUUAGGGGUACAAAAGUCUAAAGAGGAUCUUGCACCCAGUCAAGACAAGGCCCCGGC_C_UC____________________ ...........................(((((..........))))).............(.((..(((((...........)))))..)))............................ ( -9.82 = -9.52 + -0.30)
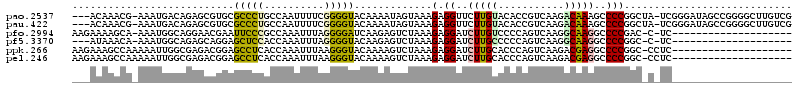

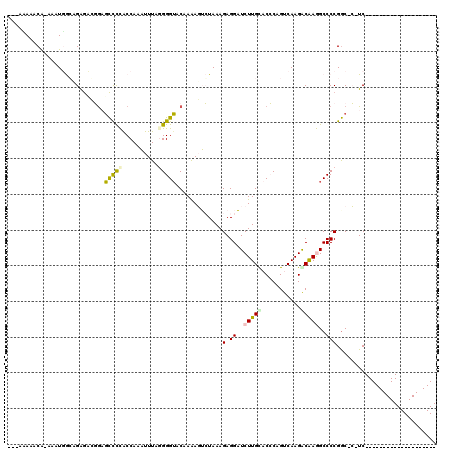
| Location | 4,772,156 – 4,772,271 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 68.78 |
| Mean single sequence MFE | -45.85 |
| Consensus MFE | -22.35 |
| Energy contribution | -22.02 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.48 |
| Mean z-score | -5.38 |
| Structure conservation index | 0.49 |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.989808 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2537 4772156 115 - 6264404 CGACAAGCCCCGGCUAUCCCGA-UAGCCGGGGCUUUGUCUUGACGGUGUACAAGAACCUCUUUACUAUUUUGUACCCCGAAAAUUGGCAGGGCGCACGCUCUGUCAUUU-CGUUUGU--- .((((((((((((((((....)-)))))))))).)))))..(((((.((((((((............)))))))).))((((..(((((((((....))))))))))))-))))...--- ( -55.70) >pau.422 5782985 115 + 6537648 CGACAAGCCCCGGCUAUCCCGA-UAGCCGGGGCUUUGUCUUGACGGUGUACAAGAACCUCUUUACUAUUUUGUACCCCGAAAAUUGGCAGGGCGCACGCUCUGUCAUUU-CGUUUGU--- .((((((((((((((((....)-)))))))))).)))))..(((((.((((((((............)))))))).))((((..(((((((((....))))))))))))-))))...--- ( -55.70) >pfo.2994 5710155 97 - 6438405 --------------------GA-G-GUCGGGGCCUUGCCUUGACUGGGGACAAGAUCCUCUUUAGACUCUUGAUCCCCUAAAUUUGGCGGGAAUUCGUUCCUGCCAUUU-UGCUUUUCUU --------------------..-(-((((((((...)))))))))((((((((((..((....))..))))).)))))..((..(((((((((....)))))))))..)-)......... ( -41.30) >pf5.3370 6367093 94 - 7074893 --------------------GA-G-GCCGGGGCCUUGCCUUGACUGGGGGCAAGAUCCUCUUUAGACUCUUGUACCCCUAAAUUUGGUGGAGCUCCUGCUCUGCCAUUU-UGUUUAU--- --------------------((-(-((....))))).........((((((((((..((....))..)))))).))))(((((.(((..((((....))))..)))...-.))))).--- ( -36.90) >ppk.266 5635735 99 + 6181863 --------------------GAGG-GCCGGGGCCUCGUCUUGACUGGGUGCAAGAUCCUCUUUAGACUUUUGUACCCUUAAAUUUGGUGAGGCUCCGUCUCGCCAAUUUUUGGCUUUCUU --------------------..((-(.(((((((((..(......((((((((((..((....))..))))))))))........)..))))))))).)))((((.....))))...... ( -42.74) >pel.246 5377299 99 + 5888780 --------------------GAGG-GCCGGGGCCUCGUCUUGACUGGGUGCAAGAUCCUCUUUAGACUUUUGUACCCUUAAAUUUGGUGAGGCUCCGUCUCGCCAAUUUUUGGCUUUCUU --------------------..((-(.(((((((((..(......((((((((((..((....))..))))))))))........)..))))))))).)))((((.....))))...... ( -42.74) >consensus ____________________GA_G_GCCGGGGCCUUGUCUUGACUGGGUACAAGAUCCUCUUUAGACUUUUGUACCCCUAAAUUUGGCGGGGCUCCGGCUCUGCCAUUU_UGUUUUU___ ............................(((((((((((......((((((((((((.......)).))))))))))........)))))))))))........................ (-22.35 = -22.02 + -0.33)

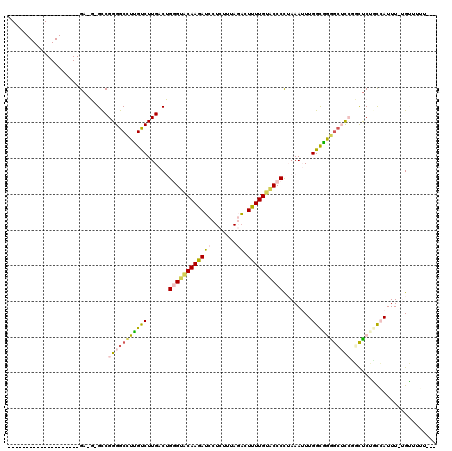
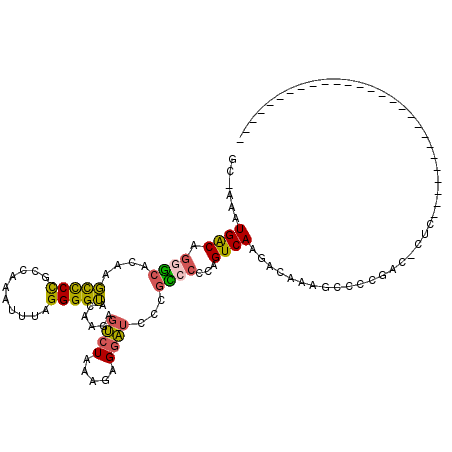
| Location | 4,772,160 – 4,772,278 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 74.45 |
| Mean single sequence MFE | -29.75 |
| Consensus MFE | -14.60 |
| Energy contribution | -14.02 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.809779 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2537 4772160 118 + 6264404 ACGAAAUGACAGAGCGUGCGCCCUGCCAAUUUUCGGGGUACAAAAUAGUAAAGAGGUUCUUGUACACCGUCAAGACAAAGCCCCGGCUAUCGGGAUAGCCGGGGCUUGUCGUUGAUGC .(((((((.(((.((....)).))).))..)))))(((((((((((.........))).)))))).))((((((((((.(((((((((((....)))))))))))))))).))))).. ( -50.80) >pst.393 5710150 89 - 6397126 GC-AAAUGACGAGGCACAAGCCCCGCCAAAUUUAGGGGUACAAGAGUCUAAAGAGGAUCCCGCCCCCAGUCAAGACAAAGCCCCGGC-CUC--------------------------- ..-.......(((((....(((((..........)))))......((((.....((........))......)))).........))-)))--------------------------- ( -21.20) >psp.2676 5231974 89 + 5928787 GC-AAAUGACAGGGCACAAGCCCCGCCAAAUUUAGGGGUACAAGAGUCUAAAGAGGAUCCCGUCCCCAGUCAAGACAAAGCCCCGAC-AUC--------------------------- ..-..(((.(.((((....(((((..........)))))......((((...(.((((...)))).).....))))...)))).).)-)).--------------------------- ( -23.00) >pfo.2994 5710162 90 + 6438405 GCAAAAUGGCAGGAACGAAUUCCCGCCAAAUUUAGGGGAUCAAGAGUCUAAAGAGGAUCUUGUCCCCAGUCAAGGCAAGGCCCCGAC-CUC--------------------------- ......((((.((((....)))).))))......(((((.(((((..((....))..)))))))))).(((..(((...)))..)))-...--------------------------- ( -32.50) >pf5.3370 6367097 90 + 7074893 ACAAAAUGGCAGAGCAGGAGCUCCACCAAAUUUAGGGGUACAAGAGUCUAAAGAGGAUCUUGCCCCCAGUCAAGGCAAGGCCCCGGC-CUC--------------------------- ......(((..((((....))))..)))......((((..(((((..((....))..))))).)))).((....)).((((....))-)).--------------------------- ( -29.30) >pss.2754 5408012 89 + 6093698 GC-AAAUGACAGGGCACAGGCCCCGCCAAAUUUAGGGGUACAAGAGUCUAAAGAGGAUCCCGCCCCCAGUCAAGACAAAGCCCCGAC-GUC--------------------------- ..-...((((.((((....(((((..........)))))....(..(((.....)))..).))))...))))...............-...--------------------------- ( -21.70) >consensus GC_AAAUGACAGGGCACAAGCCCCGCCAAAUUUAGGGGUACAAGAGUCUAAAGAGGAUCCCGCCCCCAGUCAAGACAAAGCCCCGAC_CUC___________________________ ......((((.((((....(((((..........)))))......((((.....))))...))))...)))).............................................. (-14.60 = -14.02 + -0.58)

| Location | 4,772,160 – 4,772,278 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 74.45 |
| Mean single sequence MFE | -40.74 |
| Consensus MFE | -31.31 |
| Energy contribution | -28.98 |
| Covariance contribution | -2.32 |
| Combinations/Pair | 1.56 |
| Mean z-score | -3.92 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.979889 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2537 4772160 118 - 6264404 GCAUCAACGACAAGCCCCGGCUAUCCCGAUAGCCGGGGCUUUGUCUUGACGGUGUACAAGAACCUCUUUACUAUUUUGUACCCCGAAAAUUGGCAGGGCGCACGCUCUGUCAUUUCGU ...((((.((((((((((((((((....))))))))))).))))))))).((.((((((((............))))))))))(((((..(((((((((....)))))))))))))). ( -57.90) >pst.393 5710150 89 + 6397126 ---------------------------GAG-GCCGGGGCUUUGUCUUGACUGGGGGCGGGAUCCUCUUUAGACUCUUGUACCCCUAAAUUUGGCGGGGCUUGUGCCUCGUCAUUU-GC ---------------------------(((-((..((((((((((......((((((((((..((....))..)))))).)))).......))))))))))..))))).......-.. ( -35.02) >psp.2676 5231974 89 - 5928787 ---------------------------GAU-GUCGGGGCUUUGUCUUGACUGGGGACGGGAUCCUCUUUAGACUCUUGUACCCCUAAAUUUGGCGGGGCUUGUGCCCUGUCAUUU-GC ---------------------------...-((((((((...)))))))).((((((((((..((....))..)))))).))))......(((((((((....)))))))))...-.. ( -37.70) >pfo.2994 5710162 90 - 6438405 ---------------------------GAG-GUCGGGGCCUUGCCUUGACUGGGGACAAGAUCCUCUUUAGACUCUUGAUCCCCUAAAUUUGGCGGGAAUUCGUUCCUGCCAUUUUGC ---------------------------..(-((((((((...)))))))))((((((((((..((....))..))))).)))))..((..(((((((((....)))))))))..)).. ( -41.30) >pf5.3370 6367097 90 - 7074893 ---------------------------GAG-GCCGGGGCCUUGCCUUGACUGGGGGCAAGAUCCUCUUUAGACUCUUGUACCCCUAAAUUUGGUGGAGCUCCUGCUCUGCCAUUUUGU ---------------------------(((-((....))))).........((((((((((..((....))..)))))).))))..((..(((..((((....))))..)))..)).. ( -36.30) >pss.2754 5408012 89 - 6093698 ---------------------------GAC-GUCGGGGCUUUGUCUUGACUGGGGGCGGGAUCCUCUUUAGACUCUUGUACCCCUAAAUUUGGCGGGGCCUGUGCCCUGUCAUUU-GC ---------------------------...-((((((((...)))))))).((((((((((..((....))..)))))).))))......(((((((((....)))))))))...-.. ( -36.20) >consensus ___________________________GAG_GCCGGGGCUUUGUCUUGACUGGGGACAAGAUCCUCUUUAGACUCUUGUACCCCUAAAUUUGGCGGGGCUUGUGCCCUGUCAUUU_GC .................................((((((...))))))...((((((((((((.......)).)))))).))))......(((((((((....)))))))))...... (-31.31 = -28.98 + -2.32)


Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:10:58 2007