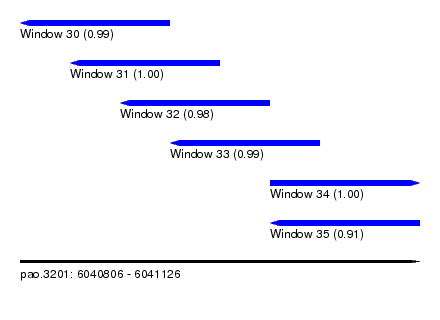
| Sequence ID | pao.3201 |
|---|---|
| Location | 6,040,806 – 6,041,126 |
| Length | 320 |
| Max. P | 0.999461 |

| Location | 6,040,806 – 6,040,926 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.67 |
| Mean single sequence MFE | -43.50 |
| Consensus MFE | -43.50 |
| Energy contribution | -43.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.33 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.31 |
| SVM RNA-class probability | 0.992188 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.3201 6040806 120 - 6264404 UGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCGCAAGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCCUU .((...(((((((...((((((..((((..(((((((((((.((....))..))))))..)))))))))))))))...)))((((((.......))))))...)))).)).......... ( -44.10) >pel.69 5769338 120 + 5888780 UGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCGCAAGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCCUU .((...(((((((...((((((..((((..(((((((((((.((....))..))))))..)))))))))))))))...)))((((((.......))))))...)))).)).......... ( -44.10) >psp.1833 3632677 120 - 5928787 UGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCGCAAGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCCUU .((...(((((((...((((((..((((..(((((((((((.((....))..))))))..)))))))))))))))...)))((((((.......))))))...)))).)).......... ( -44.10) >pf5.3378 6383963 120 - 7074893 UGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCUAACGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCCUU .((...(((((((...((((((..((((..(((((((((((.((....))..))))))..)))))))))))))))...)))((((((.......))))))...)))).)).......... ( -42.30) >pfo.3002 5727584 120 - 6438405 UGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCUAACGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCCUU .((...(((((((...((((((..((((..(((((((((((.((....))..))))))..)))))))))))))))...)))((((((.......))))))...)))).)).......... ( -42.30) >pst.2031 3870673 120 - 6397126 UGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCGCAAGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCCUU .((...(((((((...((((((..((((..(((((((((((.((....))..))))))..)))))))))))))))...)))((((((.......))))))...)))).)).......... ( -44.10) >consensus UGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCGCAAGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCCUU .((...(((((((...((((((..((((..(((((((((((.((....))..))))))..)))))))))))))))...)))((((((.......))))))...)))).)).......... (-43.50 = -43.50 + 0.00)


| Location | 6,040,846 – 6,040,966 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.67 |
| Mean single sequence MFE | -43.00 |
| Consensus MFE | -43.00 |
| Energy contribution | -43.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.82 |
| Structure conservation index | 1.00 |
| SVM decision value | 3.62 |
| SVM RNA-class probability | 0.999461 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.3201 6040846 120 - 6264404 AAACACAGCACUCUGCAAACACGAAAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCGCAAGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGG ...((((.(.((.(((...(((....)))...))).))).))))........(((.((((((..((((..(((((((((((.((....))..))))))..)))))))))))))))..))) ( -43.60) >pel.69 5769378 120 + 5888780 AAACACAGCACUCUGCAAACACGAAAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCGCAAGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGG ...((((.(.((.(((...(((....)))...))).))).))))........(((.((((((..((((..(((((((((((.((....))..))))))..)))))))))))))))..))) ( -43.60) >psp.1833 3632717 120 - 5928787 AAACACAGCACUCUGCAAACACGAAAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCGCAAGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGG ...((((.(.((.(((...(((....)))...))).))).))))........(((.((((((..((((..(((((((((((.((....))..))))))..)))))))))))))))..))) ( -43.60) >pf5.3378 6384003 120 - 7074893 AAACACAGCACUCUGCAAACACGAAAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCUAACGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGG ...((((.(.((.(((...(((....)))...))).))).))))........(((.((((((..((((..(((((((((((.((....))..))))))..)))))))))))))))..))) ( -41.80) >pfo.3002 5727624 120 - 6438405 AAACACAGCACUCUGCAAACACGAAAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCUAACGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGG ...((((.(.((.(((...(((....)))...))).))).))))........(((.((((((..((((..(((((((((((.((....))..))))))..)))))))))))))))..))) ( -41.80) >pst.2031 3870713 120 - 6397126 AAACACAGCACUCUGCAAACACGAAAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCGCAAGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGG ...((((.(.((.(((...(((....)))...))).))).))))........(((.((((((..((((..(((((((((((.((....))..))))))..)))))))))))))))..))) ( -43.60) >consensus AAACACAGCACUCUGCAAACACGAAAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCGCAAGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGG ...((((.(.((.(((...(((....)))...))).))).))))........(((.((((((..((((..(((((((((((.((....))..))))))..)))))))))))))))..))) (-43.00 = -43.00 + 0.00)


| Location | 6,040,886 – 6,041,006 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -40.80 |
| Consensus MFE | -41.02 |
| Energy contribution | -40.80 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.13 |
| Structure conservation index | 1.01 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977288 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.3201 6040886 120 - 6264404 GGCUGGUCGAAGAUACCAGGCCGCUGCGACUGUUUAUUAAAAACACAGCACUCUGCAAACACGAAAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGU ..((.(((((....(((.(((....(((..(((((.....)))))..((((((((....(((....)))......))))))))..)))..))).)))(((....)))...))))).)).. ( -39.60) >pau.3340 6313805 120 - 6537648 GGCUGGUCGAAGAUACCAGGCCGCUGCGACUGUUUAUUAAAAACACAGCACUCUGCAAACACGAAAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGU ..((.(((((....(((.(((....(((..(((((.....)))))..((((((((....(((....)))......))))))))..)))..))).)))(((....)))...))))).)).. ( -39.60) >pss.1919 3848752 120 - 6093698 UGCCGGUCGAAGAUACCAGGCCGCUGCGACUGUUUAUUAAAAACACAGCACUCUGCAAACACGAAAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGU ..((.(((((....(((.(((....(((..(((((.....)))))..((((((((....(((....)))......))))))))..)))..))).)))(((....)))...))))).)).. ( -41.40) >pf5.3378 6384043 120 - 7074893 UGCCGGUCGAAGAUACCAGGCCGCUGCGACUGUUUAUUAAAAACACAGCACUCUGCAAACACGAAAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGU ..((.(((((....(((.(((....(((..(((((.....)))))..((((((((....(((....)))......))))))))..)))..))).)))(((....)))...))))).)).. ( -41.40) >pfo.3002 5727664 120 - 6438405 CGCCGGUCGAAGAUACCAGGCCGCUGCGACUGUUUAUUAAAAACACAGCACUCUGCAAACACGAAAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGU ..((.(((((....(((.(((....(((..(((((.....)))))..((((((((....(((....)))......))))))))..)))..))).)))(((....)))...))))).)).. ( -41.40) >pst.2031 3870753 120 - 6397126 CGCCGGUCGAAGAUACCAGGCCGCUGCGACUGUUUAUUAAAAACACAGCACUCUGCAAACACGAAAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGU ..((.(((((....(((.(((....(((..(((((.....)))))..((((((((....(((....)))......))))))))..)))..))).)))(((....)))...))))).)).. ( -41.40) >consensus CGCCGGUCGAAGAUACCAGGCCGCUGCGACUGUUUAUUAAAAACACAGCACUCUGCAAACACGAAAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGU ..((.(((((....(((.(((....(((..(((((.....)))))..((((((((....(((....)))......))))))))..)))..))).)))(((....)))...))))).)).. (-41.02 = -40.80 + -0.22)

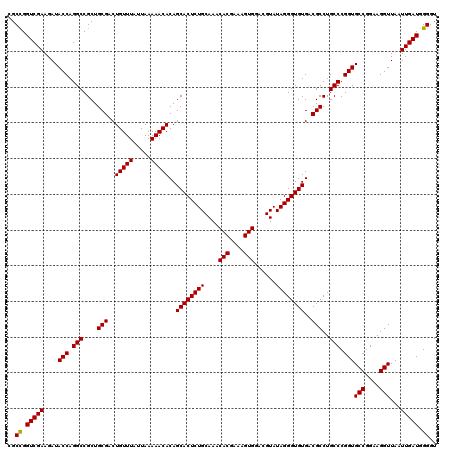
| Location | 6,040,926 – 6,041,046 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.33 |
| Mean single sequence MFE | -41.17 |
| Consensus MFE | -40.75 |
| Energy contribution | -39.17 |
| Covariance contribution | -1.58 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.985999 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.3201 6040926 120 - 6264404 GGUGCGCCGGCUAGGGUGAAGGAUUUACUCCGUAAGCUCUGGCUGGUCGAAGAUACCAGGCCGCUGCGACUGUUUAUUAAAAACACAGCACUCUGCAAACACGAAAGUGGACGUAUAGGG (((((((((((((((((...(((.....)))....))))))))))).))....))))...((((((....(((((.....))))))))).((.(((...(((....)))...))).)))) ( -42.60) >pau.3340 6313845 120 - 6537648 GGUGCGCCGGCUAGGGUGAAGGAUUUACUCCGUAAGCUCUGGCUGGUCGAAGAUACCAGGCCGCUGCGACUGUUUAUUAAAAACACAGCACUCUGCAAACACGAAAGUGGACGUAUAGGG (((((((((((((((((...(((.....)))....))))))))))).))....))))...((((((....(((((.....))))))))).((.(((...(((....)))...))).)))) ( -42.60) >pss.1919 3848792 120 - 6093698 GGUGCGCCGGUGGAGGUGAAGCAUUUACUGCGUAAGCCCCUGCCGGUCGAAGAUACCAGGCCGCUGCGACUGUUUAUUAAAAACACAGCACUCUGCAAACACGAAAGUGGACGUAUAGGG ((((((((((..(.(((...(((.....)))....))).)..)))).))....))))...((((((....(((((.....))))))))).((.(((...(((....)))...))).)))) ( -39.80) >pel.69 5769458 120 + 5888780 GGUGCGCCGGUGAGGGUGAAGCACUUGCUGCGUAAGCCCACGCCGGUCGAAGAUACCAGGCCGCUGCGACUGUUUAUUAAAAACACAGCACUCUGCAAACACGAAAGUGGACGUAUAGGG ((((((((((((.((((...(((.....)))....)))).)))))).))....))))...((((((....(((((.....))))))))).((.(((...(((....)))...))).)))) ( -42.40) >pf5.3378 6384083 120 - 7074893 GGUGCGCCGGUGAGGGUGAAGGACUUGCUCCGUAAGCUCAUGCCGGUCGAAGAUACCAGGCCGCUGCGACUGUUUAUUAAAAACACAGCACUCUGCAAACACGAAAGUGGACGUAUAGGG ((((((((((((.((((...(((.....)))....)))).)))))).))....))))...((((((....(((((.....))))))))).((.(((...(((....)))...))).)))) ( -37.20) >pfo.3002 5727704 120 - 6438405 GGUGCGCCGGUGAGGGUGAAGCACUUGCUGCGUAAGCCCACGCCGGUCGAAGAUACCAGGCCGCUGCGACUGUUUAUUAAAAACACAGCACUCUGCAAACACGAAAGUGGACGUAUAGGG ((((((((((((.((((...(((.....)))....)))).)))))).))....))))...((((((....(((((.....))))))))).((.(((...(((....)))...))).)))) ( -42.40) >consensus GGUGCGCCGGUGAGGGUGAAGCACUUACUCCGUAAGCCCACGCCGGUCGAAGAUACCAGGCCGCUGCGACUGUUUAUUAAAAACACAGCACUCUGCAAACACGAAAGUGGACGUAUAGGG ((((((((((((.((((...(((.....)))....)))).)))))).))....))))...((((((....(((((.....))))))))).((.(((...(((....)))...))).)))) (-40.75 = -39.17 + -1.58)

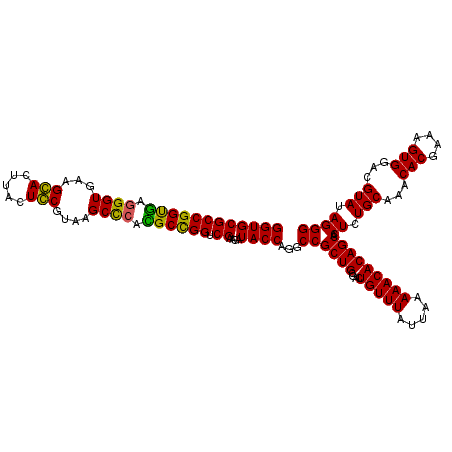
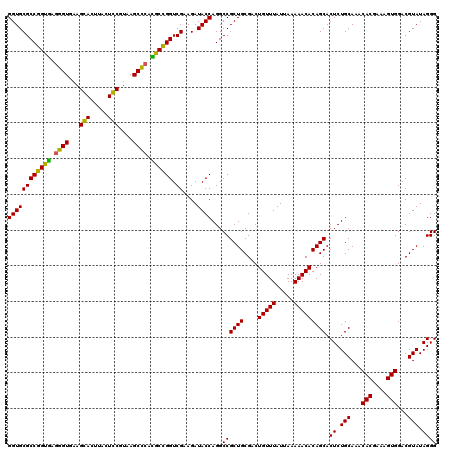
| Location | 6,041,006 – 6,041,126 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.72 |
| Mean single sequence MFE | -30.28 |
| Consensus MFE | -29.42 |
| Energy contribution | -28.67 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.63 |
| SVM RNA-class probability | 0.995880 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.3201 6041006 120 + 6264404 AGAGCUUACGGAGUAAAUCCUUCACCCUAGCCGGCGCACCUUCUCCCGAAGUUACGGUGCCAUUUUGCCUAGUUCCUUCACCCGAGUUCUCUCAAGCGCCUUGGUAUUCUCUACCCGACC ((((..((((((.....)))............(((((..........((((..(((((........)))..))..))))....(((....)))..)))))...)))..))))........ ( -30.70) >pau.3340 6313925 120 + 6537648 AGAGCUUACGGAGUAAAUCCUUCACCCUAGCCGGCGCACCUUCUCCCGAAGUUACGGUGCCAUUUUGCCUAGUUCCUUCACCCGAGUUCUCUCAAGCGCCUUGGUAUUCUCUACCCGACC ((((..((((((.....)))............(((((..........((((..(((((........)))..))..))))....(((....)))..)))))...)))..))))........ ( -30.70) >pss.1919 3848872 120 + 6093698 GGGGCUUACGCAGUAAAUGCUUCACCUCCACCGGCGCACCUUCUCCCGAAGUUACGGUGCCAUUUUGCCUAGUUCCUUCACCCGAGUUCUCUCAAGCGCCUUGGUAUUCUCUACCCAACC (((......(((.....))).........((((((((..........((((..(((((........)))..))..))))....(((....)))..)))))..)))........))).... ( -31.10) >pel.69 5769538 120 - 5888780 UGGGCUUACGCAGCAAGUGCUUCACCCUCACCGGCGCACCUUCUCCCGAAGUUACGGUGCCAUUUUGCCUAGUUCCUUCACCCGAGUUCUCUCAAGCGCCUUGGUAUUCUCUACCUAACC .(((.....(((.....)))....))).....(((((..........((((..(((((........)))..))..))))....(((....)))..)))))..((((.....))))..... ( -29.10) >pf5.3378 6384163 120 + 7074893 UGAGCUUACGGAGCAAGUCCUUCACCCUCACCGGCGCACCUUCUCCCGAAGUUACGGUGCCAUUUUGCCUAGUUCCUUCACCCGAGUUCUCUCAAGCGCCUUGGUAUUCUCUACCCAACC .(((..(((((((......)))).........(((((..........((((..(((((........)))..))..))))....(((....)))..)))))...)))..)))......... ( -29.10) >pfo.3002 5727784 120 + 6438405 UGGGCUUACGCAGCAAGUGCUUCACCCUCACCGGCGCACCUUCUCCCGAAGUUACGGUGCCAUUUUGCCUAGUUCCUUCACCCGAGUUCUCUCAAGCGCCUUGGUAUUCUCUACCCAACC ((((.....(.(((....))).)......((((((((..........((((..(((((........)))..))..))))....(((....)))..)))))..)))........))))... ( -31.00) >consensus UGAGCUUACGCAGCAAAUCCUUCACCCUCACCGGCGCACCUUCUCCCGAAGUUACGGUGCCAUUUUGCCUAGUUCCUUCACCCGAGUUCUCUCAAGCGCCUUGGUAUUCUCUACCCAACC .(((..((((((.....)))............(((((..........((((..(((((........)))..))..))))....(((....)))..)))))...)))..)))......... (-29.42 = -28.67 + -0.75)

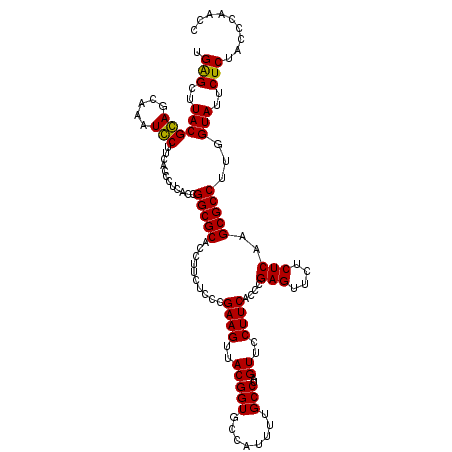
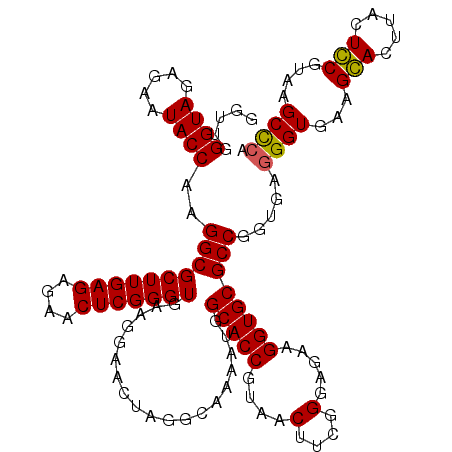
| Location | 6,041,006 – 6,041,126 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.72 |
| Mean single sequence MFE | -38.18 |
| Consensus MFE | -36.25 |
| Energy contribution | -35.67 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.907111 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.3201 6041006 120 - 6264404 GGUCGGGUAGAGAAUACCAAGGCGCUUGAGAGAACUCGGGUGAAGGAACUAGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGCUAGGGUGAAGGAUUUACUCCGUAAGCUCU ((((((((..((....((....((((((((....))))))))..))..))..))......(((((....(....).....))))).))))))(((((...(((.....)))....))))) ( -39.10) >pau.3340 6313925 120 - 6537648 GGUCGGGUAGAGAAUACCAAGGCGCUUGAGAGAACUCGGGUGAAGGAACUAGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGCUAGGGUGAAGGAUUUACUCCGUAAGCUCU ((((((((..((....((....((((((((....))))))))..))..))..))......(((((....(....).....))))).))))))(((((...(((.....)))....))))) ( -39.10) >pss.1919 3848872 120 - 6093698 GGUUGGGUAGAGAAUACCAAGGCGCUUGAGAGAACUCGGGUGAAGGAACUAGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGUGGAGGUGAAGCAUUUACUGCGUAAGCCCC ((((.((((.....))))....((((((((....))))))))....)))).(((......(((((....(....).....)))))(((((((((.(.....).))))))).))..))).. ( -37.30) >pel.69 5769538 120 + 5888780 GGUUAGGUAGAGAAUACCAAGGCGCUUGAGAGAACUCGGGUGAAGGAACUAGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGUGAGGGUGAAGCACUUGCUGCGUAAGCCCA ((((.((((.....))))....((((((((....))))))))....)))).(((......(((((....(....).....)))))(((((..((.(.....).))..))).))..))).. ( -39.20) >pf5.3378 6384163 120 - 7074893 GGUUGGGUAGAGAAUACCAAGGCGCUUGAGAGAACUCGGGUGAAGGAACUAGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGUGAGGGUGAAGGACUUGCUCCGUAAGCUCA ((((.((((.....))))....((((((((....))))))))....)))).(((......(((((....(....).....)))))))).....((((...(((.....)))....)))). ( -35.40) >pfo.3002 5727784 120 - 6438405 GGUUGGGUAGAGAAUACCAAGGCGCUUGAGAGAACUCGGGUGAAGGAACUAGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGUGAGGGUGAAGCACUUGCUGCGUAAGCCCA ((((.((((.....))))....((((((((....))))))))....)))).(((......(((((....(....).....)))))(((((..((.(.....).))..))).))..))).. ( -39.00) >consensus GGUUGGGUAGAGAAUACCAAGGCGCUUGAGAGAACUCGGGUGAAGGAACUAGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGUGAGGGUGAAGCACUUACUCCGUAAGCCCA .....((((.....))))..((((((((((....)))))))...................(((((....(....).....)))))))).....((((...(((.....)))....)))). (-36.25 = -35.67 + -0.58)

Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:03:51 2007