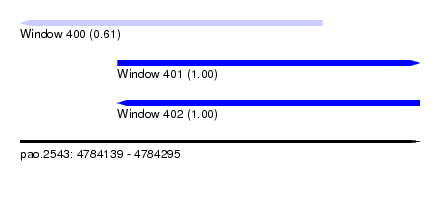
| Sequence ID | pao.2543 |
|---|---|
| Location | 4,784,139 – 4,784,295 |
| Length | 156 |
| Max. P | 0.999970 |

| Location | 4,784,139 – 4,784,257 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.95 |
| Mean single sequence MFE | -37.35 |
| Consensus MFE | -32.52 |
| Energy contribution | -32.07 |
| Covariance contribution | -0.46 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.611862 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2543 4784139 118 - 6264404 CCAGUAGCUCAAUUGGCAGAGCGGCGGUCUCCAAAACCGCAGGUUGGGGGUUCGAUUCCCUCCUGGCCUGCCAGAUUUCCCAAGUAAA-UCUGGCUUU-UCUUCACGAGAUUCCUAACAG ......((((........))))((.((((((.........((((..((((.........))))..))))(((((((((.......)))-))))))...-.......))))))))...... ( -38.40) >pau.416 5794970 116 + 6537648 CCAGUAGCUCAAUUGGCAGAGCGGCGGUCUCCAAAACCGCAGGUUGGGGGUUCGAUUCCCUCCUGGCCUGCCAGAUUUCCCAAGUAAA-UCUGGCUUU-UCUUCACGAGAUUCCUAAC-- ......((((........))))((.((((((.........((((..((((.........))))..))))(((((((((.......)))-))))))...-.......))))))))....-- ( -38.40) >pfo.3000 5722192 113 - 6438405 UCAGUAGCUCAAUUGGCAGAGCGACGGUCUCCAAAACCGUAGGUUGGGGGUUCGAUUCCCUCCUGACCUGCCAGAUUCACUCGGUGUG-UCUGGCUUUCUUUUCACAGGAUCUU------ ...((.((((........)))).))(((((..........((((..((((.........))))..))))(((((((.(((...))).)-))))))............)))))..------ ( -37.40) >pf5.3376 6379133 113 - 7074893 UCAGUAGCUCAAUUGGCAGAGCGACGGUCUCCAAAACCGUAGGUUGGGGGUUCGAUUCCCUCCUGACCUGCCAGAUUCACUCAAUGUG-UCUGGCUUUCUUUUCACAGGAUCUU------ ...((.((((........)))).))(((((..........((((..((((.........))))..))))((((((..(((.....)))-))))))............)))))..------ ( -35.90) >ppk.260 5647767 112 + 6181863 UCAGUAGCUCAAUUGGCAGAGCGACGGUCUCCAAAACCGUAGGUUGGGGGUUCGAUUCCCUCCUGACCUGCCAUUUCACCUCGGUGUGA--UGGCUUUCUUCUUACAGGAUCCU------ ...((.((((........)))).))((..(((........((((..((((.........))))..))))((((((.(((....))).))--))))............))).)).------ ( -38.00) >pel.240 5389335 112 + 5888780 UCAGUAGCUCAAUUGGCAGAGCGACGGUCUCCAAAACCGUAGGUUGGGGGUUCGAUUCCCUCCUGACCUGCCA-UUCACCUCGGUGUGAU-UGGCUUUCUUCUCACAGGAUCCU------ ...((.((((........)))).))((..(((........((((..((((.........))))..))))((((-.((((......)))).-))))............))).)).------ ( -36.00) >consensus UCAGUAGCUCAAUUGGCAGAGCGACGGUCUCCAAAACCGUAGGUUGGGGGUUCGAUUCCCUCCUGACCUGCCAGAUUACCUCAGUGUG_UCUGGCUUUCUUUUCACAGGAUCCU______ ......((((........)))).(((((.......)))))((((..((((.........))))..))))((((((.((........)).))))))......................... (-32.52 = -32.07 + -0.46)

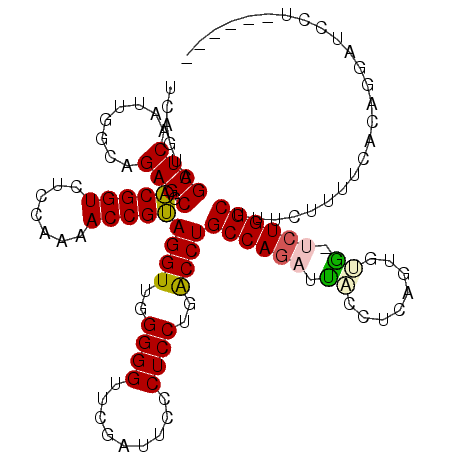
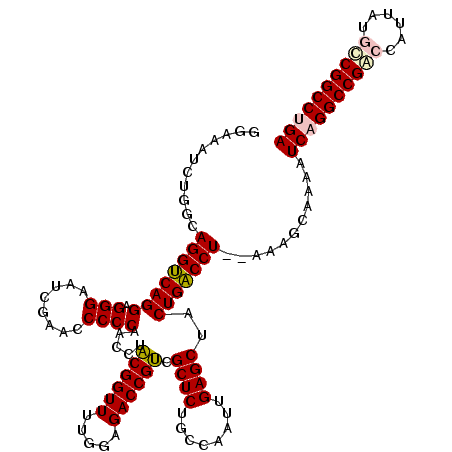
| Location | 4,784,177 – 4,784,295 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.28 |
| Mean single sequence MFE | -47.03 |
| Consensus MFE | -43.85 |
| Energy contribution | -43.80 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.93 |
| SVM decision value | 3.50 |
| SVM RNA-class probability | 0.999300 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2543 4784177 118 + 6264404 GGAAAUCUGGCAGGCCAGGAGGGAAUCGAACCCCCAACCUGCGGUUUUGGAGACCGCCGCUCUGCCAAUUGAGCUACUGGCCU--GAAGCAAAAUCAGGCCGACCAUUAUGCCGGCCUGA ......((..(((((((((.(((........)))).....((((((.....)))))).((((........))))..)))))))--).)).....((((((((..........)))))))) ( -48.30) >pau.416 5795006 118 - 6537648 GGAAAUCUGGCAGGCCAGGAGGGAAUCGAACCCCCAACCUGCGGUUUUGGAGACCGCCGCUCUGCCAAUUGAGCUACUGGCCU--GAAGCAAAAUCAGGCCGGCCAUUAUGCCGGCCUGA ......((..(((((((((.(((........)))).....((((((.....)))))).((((........))))..)))))))--).)).....((((((((((......)))))))))) ( -55.40) >pfo.3000 5722225 118 + 6438405 GUGAAUCUGGCAGGUCAGGAGGGAAUCGAACCCCCAACCUACGGUUUUGGAGACCGUCGCUCUGCCAAUUGAGCUACUGACCU--AAAACAAAAUCAGGCCGACCAUUAUGCCGGCCUGA ...........(((((((((.((..((.((..((........))..)).))..)).))((((........))))..)))))))--.........((((((((..........)))))))) ( -41.30) >pf5.3376 6379166 119 + 7074893 GUGAAUCUGGCAGGUCAGGAGGGAAUCGAACCCCCAACCUACGGUUUUGGAGACCGUCGCUCUGCCAAUUGAGCUACUGACCUA-AAAACAAAAUCAGGCCGACCAUUAUGUCGGCCCGA ...........(((((((((.((..((.((..((........))..)).))..)).))((((........))))..))))))).-.........((.(((((((......))))))).)) ( -41.70) >ppk.260 5647799 120 - 6181863 GGUGAAAUGGCAGGUCAGGAGGGAAUCGAACCCCCAACCUACGGUUUUGGAGACCGUCGCUCUGCCAAUUGAGCUACUGACCUGUAACGCUGUAUCAGGCCGGCCAUUAUACCGGCCUGA ((((.....(((((((((((.((..((.((..((........))..)).))..)).))((((........))))..)))))))))..))))...(((((((((........))))))))) ( -52.10) >pel.240 5389368 119 - 5888780 GGUGAA-UGGCAGGUCAGGAGGGAAUCGAACCCCCAACCUACGGUUUUGGAGACCGUCGCUCUGCCAAUUGAGCUACUGACCUAUAACGCUGUAUCAGGCCGGCCAUUAUACCGGCCCGA ..((((-(((((((((((((.((..((.((..((........))..)).))..)).))((((........))))..))))))).....))))).)))((((((........))))))... ( -43.40) >consensus GGAAAUCUGGCAGGUCAGGAGGGAAUCGAACCCCCAACCUACGGUUUUGGAGACCGUCGCUCUGCCAAUUGAGCUACUGACCU__AAAGCAAAAUCAGGCCGACCAUUAUGCCGGCCUGA ...........((((((((.(((........)))).....((((((.....)))))).((((........))))..)))))))...........((((((((((......)))))))))) (-43.85 = -43.80 + -0.05)
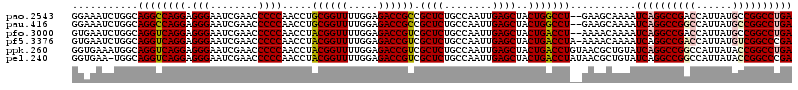
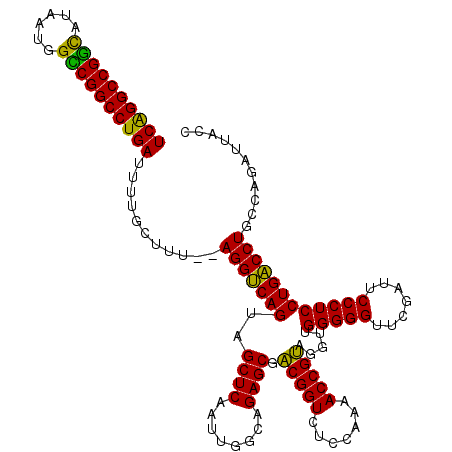
| Location | 4,784,177 – 4,784,295 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.28 |
| Mean single sequence MFE | -51.13 |
| Consensus MFE | -50.65 |
| Energy contribution | -48.93 |
| Covariance contribution | -1.72 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.21 |
| Structure conservation index | 0.99 |
| SVM decision value | 5.04 |
| SVM RNA-class probability | 0.999970 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2543 4784177 118 - 6264404 UCAGGCCGGCAUAAUGGUCGGCCUGAUUUUGCUUC--AGGCCAGUAGCUCAAUUGGCAGAGCGGCGGUCUCCAAAACCGCAGGUUGGGGGUUCGAUUCCCUCCUGGCCUGCCAGAUUUCC ((((((((((......)))))))))).((((...(--(((((((..((((........)))).(((((.......))))).....(((((.......))))))))))))).))))..... ( -54.60) >pau.416 5795006 118 + 6537648 UCAGGCCGGCAUAAUGGCCGGCCUGAUUUUGCUUC--AGGCCAGUAGCUCAAUUGGCAGAGCGGCGGUCUCCAAAACCGCAGGUUGGGGGUUCGAUUCCCUCCUGGCCUGCCAGAUUUCC ((((((((((......)))))))))).((((...(--(((((((..((((........)))).(((((.......))))).....(((((.......))))))))))))).))))..... ( -57.30) >pfo.3000 5722225 118 - 6438405 UCAGGCCGGCAUAAUGGUCGGCCUGAUUUUGUUUU--AGGUCAGUAGCUCAAUUGGCAGAGCGACGGUCUCCAAAACCGUAGGUUGGGGGUUCGAUUCCCUCCUGACCUGCCAGAUUCAC ((((((((((......)))))))))).((((...(--(((((((..((((........)))).(((((.......))))).....(((((.......))))))))))))).))))..... ( -48.40) >pf5.3376 6379166 119 - 7074893 UCGGGCCGACAUAAUGGUCGGCCUGAUUUUGUUUU-UAGGUCAGUAGCUCAAUUGGCAGAGCGACGGUCUCCAAAACCGUAGGUUGGGGGUUCGAUUCCCUCCUGACCUGCCAGAUUCAC ((((((((((......))))))))))....(((((-.((..(.((.((((........)))).)))..))..))))).((((((..((((.........))))..))))))......... ( -47.90) >ppk.260 5647799 120 + 6181863 UCAGGCCGGUAUAAUGGCCGGCCUGAUACAGCGUUACAGGUCAGUAGCUCAAUUGGCAGAGCGACGGUCUCCAAAACCGUAGGUUGGGGGUUCGAUUCCCUCCUGACCUGCCAUUUCACC ((((((((((......))))))))))..........((((((((..((((........)))).(((((.......))))).....(((((.......))))))))))))).......... ( -50.50) >pel.240 5389368 119 + 5888780 UCGGGCCGGUAUAAUGGCCGGCCUGAUACAGCGUUAUAGGUCAGUAGCUCAAUUGGCAGAGCGACGGUCUCCAAAACCGUAGGUUGGGGGUUCGAUUCCCUCCUGACCUGCCA-UUCACC ((((((((((......))))))))))..........((((((((..((((........)))).(((((.......))))).....(((((.......)))))))))))))...-...... ( -48.10) >consensus UCAGGCCGGCAUAAUGGCCGGCCUGAUUUUGCUUU__AGGUCAGUAGCUCAAUUGGCAGAGCGACGGUCUCCAAAACCGUAGGUUGGGGGUUCGAUUCCCUCCUGACCUGCCAGAUUACC ((((((((((......))))))))))...........(((((((..((((........)))).(((((.......))))).....(((((.......))))))))))))........... (-50.65 = -48.93 + -1.72)

Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:10:38 2007