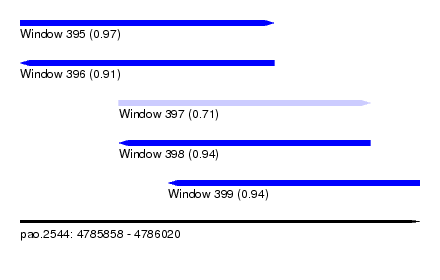
| Sequence ID | pao.2544 |
|---|---|
| Location | 4,785,858 – 4,786,020 |
| Length | 162 |
| Max. P | 0.969353 |

| Location | 4,785,858 – 4,785,961 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 74.72 |
| Mean single sequence MFE | -27.52 |
| Consensus MFE | -9.48 |
| Energy contribution | -10.03 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.34 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969353 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
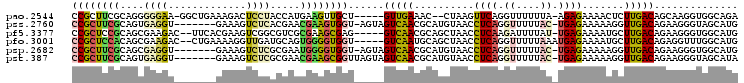
>pao.2544 4785858 103 + 6264404 CUCGGGAAUCCCUCCAAACUGGUGGGCGGCAUUCUCUACGUCUGCCACCUUGCUGUCAAGAGUUUUCUCU-UAAAAAACCUGAACUUAG--GUUUCAAC-----AGCAACU ...((....(((.((.....)).))).((((..(.....)..)))).))(((((((.(((((....))))-)...(((((((....)))--))))..))-----))))).. ( -31.80) >pau.415 5796687 103 - 6537648 CUCGGGAAUCCCUCCAAACUGGUGGGCGGCAUUCUCUACGUCUGCCACCUUGCUGUCAAGAGUUUUCUCU-UAAAAAACCUGAACUUAG--GUUUCAAC-----AGCAACU ...((....(((.((.....)).))).((((..(.....)..)))).))(((((((.(((((....))))-)...(((((((....)))--))))..))-----))))).. ( -31.80) >pf5.3377 6380851 102 + 7074893 CUCGGGAACCCCUCCUAA--GCGAGGCGACAUUCUACAU-CAUGCCACCCUUCUGUCAAGCAUUUUCUCA-AUAAAAUCUUGAGGUUAGCUGCGUUGAC-----CUCGCUU ...((((.....))))((--((((((((((.........-..((.((......)).))(((.(..(((((-(.......))))))..))))..)))).)-----))))))) ( -27.50) >pfo.3001 5723911 103 + 6438405 CUCGGGAACCCCUCCUAA--UCAGGCCGGCAUUCUAUAC-CAUGCCAAACCUCUGUCAAGCAUUUUCUCAUUUAAAAACCUGAGGUUAGCUGCAUUGAC-----ACCACCC ...(((............--..(((..(((((.......-.)))))...))).(((((((((...(((((..........))))).....))).)))))-----)...))) ( -22.80) >psp.2682 5244545 107 + 5928787 CUCGGGAACCCCUCCUAA--GCGAGGCAGCAUUCUCAACUCAUGCCACCCUUCUGUCAACCUUUUUCUCA-GUAAAAACCUGAGGUUACAUGCGUUGACUACU-ACCACCC ...(((....((((....--..))))..((((.........))))..)))....(((((((....(((((-(.......))))))......).))))))....-....... ( -23.40) >pst.387 5722721 108 - 6397126 CUCGGGAACCCCUCCUAA--GCGAGGCAGCAUUCUCAACUUAUGCUACCCUUCUGUCAACCUUUUUCUCA-GUAAAAACCUGAGGUUACAUGCGUUGACUACUAACCGCUU ...((((.....))))((--(((.((.(((((.........))))).)).....(((((((....(((((-(.......))))))......).)))))).......))))) ( -27.80) >consensus CUCGGGAACCCCUCCUAA__GCGAGGCGGCAUUCUCUAC_CAUGCCACCCUUCUGUCAAGCAUUUUCUCA_UUAAAAACCUGAGGUUAG_UGCGUUGAC_____ACCACCU ((((((....(((..........))).(((((.........)))))................................))))))........................... ( -9.48 = -10.03 + 0.56)

| Location | 4,785,858 – 4,785,961 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 74.72 |
| Mean single sequence MFE | -33.75 |
| Consensus MFE | -15.82 |
| Energy contribution | -14.88 |
| Covariance contribution | -0.94 |
| Combinations/Pair | 1.37 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.47 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.906205 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2544 4785858 103 - 6264404 AGUUGCU-----GUUGAAAC--CUAAGUUCAGGUUUUUUA-AGAGAAAACUCUUGACAGCAAGGUGGCAGACGUAGAGAAUGCCGCCCACCAGUUUGGAGGGAUUCCCGAG ..(((((-----((((((((--((......)))))))..(-((((....)))))))))))))(((((((..(.....)..)))))))..((.....)).(((...)))... ( -35.80) >pau.415 5796687 103 + 6537648 AGUUGCU-----GUUGAAAC--CUAAGUUCAGGUUUUUUA-AGAGAAAACUCUUGACAGCAAGGUGGCAGACGUAGAGAAUGCCGCCCACCAGUUUGGAGGGAUUCCCGAG ..(((((-----((((((((--((......)))))))..(-((((....)))))))))))))(((((((..(.....)..)))))))..((.....)).(((...)))... ( -35.80) >pf5.3377 6380851 102 - 7074893 AAGCGAG-----GUCAACGCAGCUAACCUCAAGAUUUUAU-UGAGAAAAUGCUUGACAGAAGGGUGGCAUG-AUGUAGAAUGUCGCCUCGC--UUAGGAGGGGUUCCCGAG ((((((.-----(((((.(((......(((((.......)-))))....)))))))).....((((((((.-.......))))))))))))--))....(((...)))... ( -34.10) >pfo.3001 5723911 103 - 6438405 GGGUGGU-----GUCAAUGCAGCUAACCUCAGGUUUUUAAAUGAGAAAAUGCUUGACAGAGGUUUGGCAUG-GUAUAGAAUGCCGGCCUGA--UUAGGAGGGGUUCCCGAG (((...(-----(((((.(((......((((..(....)..))))....))))))))).(((((.(((((.-.......))))))))))..--............)))... ( -29.10) >psp.2682 5244545 107 - 5928787 GGGUGGU-AGUAGUCAACGCAUGUAACCUCAGGUUUUUAC-UGAGAAAAAGGUUGACAGAAGGGUGGCAUGAGUUGAGAAUGCUGCCUCGC--UUAGGAGGGGUUCCCGAG (.(.(((-((((.(((((.(((((.((((...(((...((-(........))).)))....)))).))))).)))))...))))))).).)--......(((...)))... ( -34.80) >pst.387 5722721 108 + 6397126 AAGCGGUUAGUAGUCAACGCAUGUAACCUCAGGUUUUUAC-UGAGAAAAAGGUUGACAGAAGGGUAGCAUAAGUUGAGAAUGCUGCCUCGC--UUAGGAGGGGUUCCCGAG (((((.......((((((.(.......(((((.......)-)))).....)))))))....(((((((((.........))))))))))))--))....(((...)))... ( -32.90) >consensus AGGUGGU_____GUCAACGCA_CUAACCUCAGGUUUUUAA_UGAGAAAAAGCUUGACAGAAGGGUGGCAUA_GUAGAGAAUGCCGCCCCGC__UUAGGAGGGGUUCCCGAG ............(((((..........(((............))).......))))).....((((((((.........))))))))............(((...)))... (-15.82 = -14.88 + -0.94)

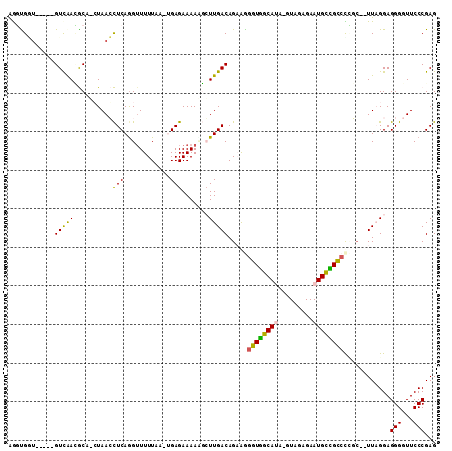
| Location | 4,785,898 – 4,786,000 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 69.72 |
| Mean single sequence MFE | -29.00 |
| Consensus MFE | -4.89 |
| Energy contribution | -5.59 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.17 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.707437 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2544 4785898 102 + 6264404 UCUGCCACCUUGCUGUCAAGAGUUUUCUCU-UAAAAAACCUGAACUUAG--GUUUCAAC-----AGCAACUUCAUGGUAGGAGUCUUUCAGCC-UUCCCCCUGCGAAGCGG (((((((..(((((((.(((((....))))-)...(((((((....)))--))))..))-----))))).....))))))).(.(((((((..-......))).))))).. ( -27.50) >pss.2760 5420623 102 + 6093698 CAUGCUACCCUUCUGUCAACCUUUUUCUCA-GUAAAAACCUGAGGUUACAUGCGUUGACUACU-ACCACUUCGUUCGUGAGACUUUC-------ACCUCACUGCGAAGCGG ..............(((((((....(((((-(.......))))))......).))))))....-.((.((((((..(((((......-------..))))).)))))).)) ( -29.70) >pf5.3377 6380888 103 + 7074893 CAUGCCACCCUUCUGUCAAGCAUUUUCUCA-AUAAAAUCUUGAGGUUAGCUGCGUUGAC-----CUCGCUUCGCGACGCCGACUUCGUGAA--GUCUUCGCUGCGGAGCGG ..............((((((((...(((((-(.......)))))).....))).)))))-----..(((((((((.((..(((((....))--)))..)).))))))))). ( -34.20) >pfo.3001 5723948 104 + 6438405 CAUGCCAAACCUCUGUCAAGCAUUUUCUCAUUUAAAAACCUGAGGUUAGCUGCAUUGAC-----ACCACCCCACUGCAUCAACCUUUUCAG--GUCUUCGCUGUGGAGCGG .............(((((((((...(((((..........))))).....))).)))))-----)((.(.((((.((....((((....))--))....)).)))).).)) ( -25.10) >psp.2682 5244583 102 + 5928787 CAUGCCACCCUUCUGUCAACCUUUUUCUCA-GUAAAAACCUGAGGUUACAUGCGUUGACUACU-ACCACCCCAUUCGCGAGACUUUC-------ACCUCGCUGCGAAGCGG .......((((((.(((((((....(((((-(.......))))))......).))))))....-............(((((......-------..)))))...)))).)) ( -26.30) >pst.387 5722759 103 - 6397126 UAUGCUACCCUUCUGUCAACCUUUUUCUCA-GUAAAAACCUGAGGUUACAUGCGUUGACUACUAACCGCUUCGUUCGCGAGACUUUC-------ACCUCACUGCGAAGCGG ..............(((((((....(((((-(.......))))))......).))))))......(((((((((..(.(((......-------..))).).))))))))) ( -31.20) >consensus CAUGCCACCCUUCUGUCAACCUUUUUCUCA_GUAAAAACCUGAGGUUACAUGCGUUGAC_____ACCACCUCGUUCGCGAGACUUUC_______ACCUCGCUGCGAAGCGG ..............((((((.....((((............))))........)))))).........................................((((...)))) ( -4.89 = -5.59 + 0.69)


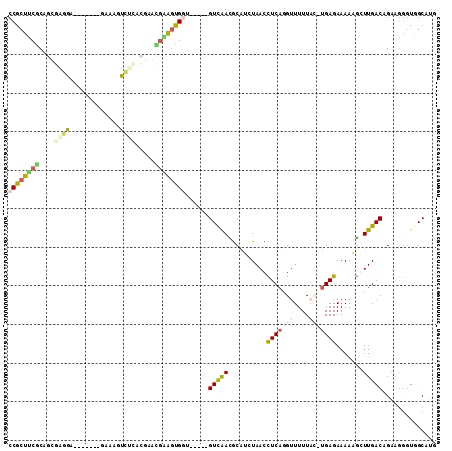
| Location | 4,785,898 – 4,786,000 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 69.72 |
| Mean single sequence MFE | -34.09 |
| Consensus MFE | -10.68 |
| Energy contribution | -11.22 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.61 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.31 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.939020 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2544 4785898 102 - 6264404 CCGCUUCGCAGGGGGAA-GGCUGAAAGACUCCUACCAUGAAGUUGCU-----GUUGAAAC--CUAAGUUCAGGUUUUUUA-AGAGAAAACUCUUGACAGCAAGGUGGCAGA (((((....(((((...-..........))))).........(((((-----((((((((--((......)))))))..(-((((....)))))))))))))))))).... ( -29.32) >pss.2760 5420623 102 - 6093698 CCGCUUCGCAGUGAGGU-------GAAAGUCUCACGAACGAAGUGGU-AGUAGUCAACGCAUGUAACCUCAGGUUUUUAC-UGAGAAAAAGGUUGACAGAAGGGUAGCAUG ((((((((..(((((((-------....)))))))...)))))))).-....((((((.(.......(((((.......)-)))).....))))))).............. ( -33.30) >pf5.3377 6380888 103 - 7074893 CCGCUCCGCAGCGAAGAC--UUCACGAAGUCGGCGUCGCGAAGCGAG-----GUCAACGCAGCUAACCUCAAGAUUUUAU-UGAGAAAAUGCUUGACAGAAGGGUGGCAUG (((((((((.((((.(((--((....)))))....))))...)))..-----(((((.(((......(((((.......)-))))....))))))))....)))))).... ( -35.50) >pfo.3001 5723948 104 - 6438405 CCGCUCCACAGCGAAGAC--CUGAAAAGGUUGAUGCAGUGGGGUGGU-----GUCAAUGCAGCUAACCUCAGGUUUUUAAAUGAGAAAAUGCUUGACAGAGGUUUGGCAUG (((((((((.(((..(((--((....)))))..))).))))))))).-----....((((....((((((..(((.....((......))....))).))))))..)))). ( -39.80) >psp.2682 5244583 102 - 5928787 CCGCUUCGCAGCGAGGU-------GAAAGUCUCGCGAAUGGGGUGGU-AGUAGUCAACGCAUGUAACCUCAGGUUUUUAC-UGAGAAAAAGGUUGACAGAAGGGUGGCAUG ((((((((..(((((((-------....)))))))...)))))))).-....((((((.(.......(((((.......)-)))).....))))))).............. ( -31.80) >pst.387 5722759 103 + 6397126 CCGCUUCGCAGUGAGGU-------GAAAGUCUCGCGAACGAAGCGGUUAGUAGUCAACGCAUGUAACCUCAGGUUUUUAC-UGAGAAAAAGGUUGACAGAAGGGUAGCAUA ((((((((..(((((((-------....)))))))...))))))))......((((((.(.......(((((.......)-)))).....))))))).............. ( -34.80) >consensus CCGCUUCGCAGCGAGGA_______GAAAGUCUCACGAACGAAGUGGU_____GUCAACGCAUCUAACCUCAGGUUUUUAC_UGAGAAAAAGCUUGACAGAAGGGUGGCAUG ((((((((....((((.............)))).....))))))))......(((((..........((((.((....)).)))).......))))).............. (-10.68 = -11.22 + 0.53)

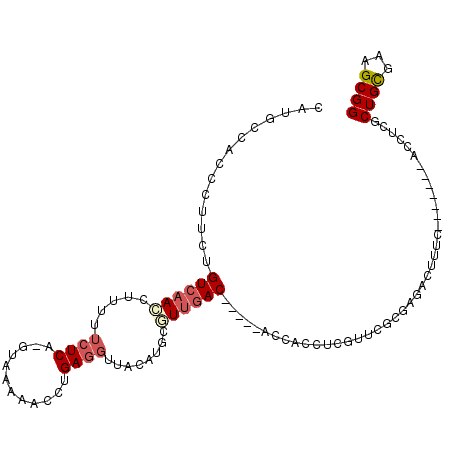
| Location | 4,785,918 – 4,786,020 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 69.71 |
| Mean single sequence MFE | -33.13 |
| Consensus MFE | -10.10 |
| Energy contribution | -10.64 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.64 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.30 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.936421 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2544 4785918 102 - 6264404 UGGUUUGCAUAGAAUGGCGCCCGCUUCGCAGGGGGAA-GGCUGAAAGACUCCUACCAUGAAGUUGCU-----GUUGAAAC--CUAAGUUCAGGUUUUUUA-AGAGAAAACU .(((((.(....(((((((...((((((..((((((.-(.........))))).)).))))))))))-----)))(((((--((......)))))))...-...).))))) ( -22.20) >pss.2760 5420643 99 - 6093698 ---UUUACAUAGAAUGGCGCCCGCUUCGCAGUGAGGU-------GAAAGUCUCACGAACGAAGUGGU-AGUAGUCAACGCAUGUAACCUCAGGUUUUUAC-UGAGAAAAAG ---.((((((.(..((((..((((((((..(((((((-------....)))))))...)))))))).-....))))...))))))).(((((.......)-))))...... ( -34.30) >pf5.3377 6380908 101 - 7074893 --GGUUGCAUAGAAUGGCGCCCGCUCCGCAGCGAAGAC--UUCACGAAGUCGGCGUCGCGAAGCGAG-----GUCAACGCAGCUAACCUCAAGAUUUUAU-UGAGAAAAUG --((((((......((((.(.((((.(((.(((..(((--((....)))))..))).))).)))).)-----))))..))))))...(((((.......)-))))...... ( -35.40) >pfo.3001 5723968 100 - 6438405 ----UUGCAUAGAAUGGCGCCCGCUCCACAGCGAAGAC--CUGAAAAGGUUGAUGCAGUGGGGUGGU-----GUCAAUGCAGCUAACCUCAGGUUUUUAAAUGAGAAAAUG ----((((((....(((((((.(((((((.(((..(((--((....)))))..))).))))))))))-----)))))))))).....((((..(....)..))))...... ( -40.40) >psp.2682 5244603 97 - 5928787 -----UACAUAGAAUGGCGCCCGCUUCGCAGCGAGGU-------GAAAGUCUCGCGAAUGGGGUGGU-AGUAGUCAACGCAUGUAACCUCAGGUUUUUAC-UGAGAAAAAG -----(((((.(..((((..((((((((..(((((((-------....)))))))...)))))))).-....))))...))))))..(((((.......)-))))...... ( -32.00) >pst.387 5722779 98 + 6397126 -----UACAUAGAAUGGCGCCCGCUUCGCAGUGAGGU-------GAAAGUCUCGCGAACGAAGCGGUUAGUAGUCAACGCAUGUAACCUCAGGUUUUUAC-UGAGAAAAAG -----(((((.(..((((((((((((((..(((((((-------....)))))))...))))))))...)).))))...))))))..(((((.......)-))))...... ( -34.50) >consensus ____UUACAUAGAAUGGCGCCCGCUUCGCAGCGAGGA_______GAAAGUCUCACGAACGAAGUGGU_____GUCAACGCAUCUAACCUCAGGUUUUUAC_UGAGAAAAAG ..............((((..((((((((....((((.............)))).....))))))))......))))...........((((.((....)).))))...... (-10.10 = -10.64 + 0.53)

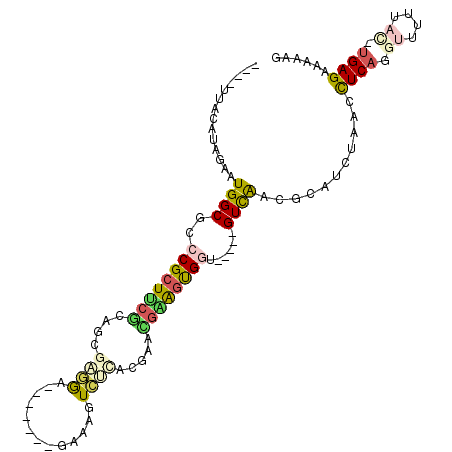
Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:10:35 2007