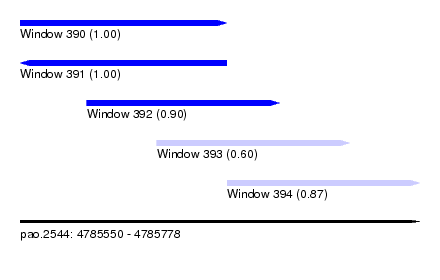
| Sequence ID | pao.2544 |
|---|---|
| Location | 4,785,550 – 4,785,778 |
| Length | 228 |
| Max. P | 0.999946 |

| Location | 4,785,550 – 4,785,668 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.49 |
| Mean single sequence MFE | -43.20 |
| Consensus MFE | -41.24 |
| Energy contribution | -40.52 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.06 |
| Mean z-score | -4.64 |
| Structure conservation index | 0.95 |
| SVM decision value | 4.75 |
| SVM RNA-class probability | 0.999946 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
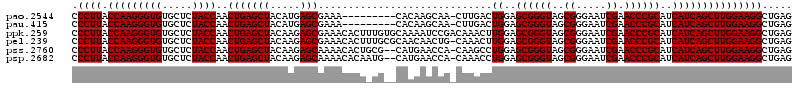
>pao.2544 4785550 118 + 6264404 AU--UAGAACAAGGGCAGAUAUUUUCAUAUCUGCCCCUGGAUAAAUGGAGCUCAUGAGCGGAUUUGAACCGCUGACCUCACCCUUACCAAGGGUGUGCUCUACCAACUGAGCUACAUGAG .(--((...((.((((((((((....)))))))))).))..)))(((.((((((..(((((.......)))))((.(.((((((.....)))))).).)).......)))))).)))... ( -45.40) >pau.415 5796379 118 - 6537648 AU--UAGAACAAGGGCAGAUAUUUUCAUAUCUGCCCCUGGAUAAAUGGAGCUCAUGAGCGGAUUUGAACCGCUGACCUCACCCUUACCAAGGGUGUGCUCUACCAACUGAGCUACAUGAG .(--((...((.((((((((((....)))))))))).))..)))(((.((((((..(((((.......)))))((.(.((((((.....)))))).).)).......)))))).)))... ( -45.40) >ppk.259 5649165 118 - 6181863 -UGAUAAAACAAAGGCAGAUAUUUUCAUAUCUGCCUU-GUUUAUAUGGAGCUCUUGAGCGGACUUGAACCGCUGACCUCACCCUUACCAAGGGUGUGCUCUACCAACUGAGCUACAAGAG -..(((((...(((((((((((....)))))))))))-.)))))......(((((((((((.......))))).....((((((.....)))))).((((........))))..)))))) ( -44.50) >pel.239 5390734 118 - 5888780 -UGAUAAAACAAAGGCAGAUAUUUUCAUAUCUGCCUU-GUUUAUAUGGAGCUCUUGAGCGGAUUUGAACCGCUGACCUCACCCUUACCAAGGGUGUGCUCUACCAACUGAGCUACAAGAG -..(((((...(((((((((((....)))))))))))-.)))))......(((((((((((.......))))).....((((((.....)))))).((((........))))..)))))) ( -44.50) >psp.2682 5244236 114 + 5928787 ----UAAAACAAAGGCAGAUAUUUUCAUAUCUUCCUU--UAGCAAUGGAGCUCUUGAGCGGAUUUGAACCGCUGACCUCACCCUUACCAAGGGUGUGCUCUACCAACUGAGCUACAAGAG ----......(((((.((((((....)))))).))))--)..........(((((((((((.......))))).....((((((.....)))))).((((........))))..)))))) ( -36.40) >pst.387 5722412 114 - 6397126 ----UAAAACAAAGGCAGAUAUUUUCAUAUCUGCCUU--UAGCAAUGGAGCUCUUGAGCGGAUUCGAACCGCUGACCUCACCCUUACCAAGGGUGUGCUCUACCAACUGAGCUACAAGAG ----......((((((((((((....)))))))))))--)..........(((((((((((.......))))).....((((((.....)))))).((((........))))..)))))) ( -43.00) >consensus _U__UAAAACAAAGGCAGAUAUUUUCAUAUCUGCCUU_GUAUAAAUGGAGCUCUUGAGCGGAUUUGAACCGCUGACCUCACCCUUACCAAGGGUGUGCUCUACCAACUGAGCUACAAGAG ............((((((((((....))))))))))..............(((((((((((.......))))).....((((((.....)))))).((((........))))..)))))) (-41.24 = -40.52 + -0.72)

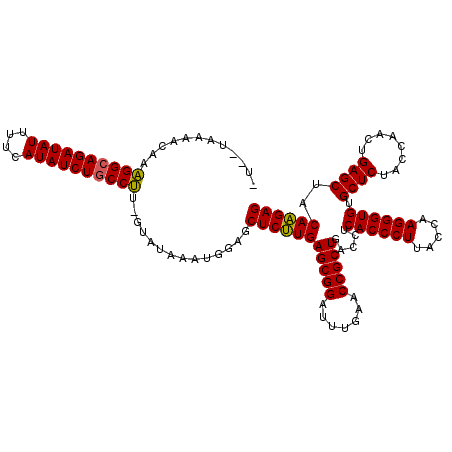
| Location | 4,785,550 – 4,785,668 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.49 |
| Mean single sequence MFE | -47.80 |
| Consensus MFE | -45.09 |
| Energy contribution | -44.15 |
| Covariance contribution | -0.94 |
| Combinations/Pair | 1.09 |
| Mean z-score | -5.15 |
| Structure conservation index | 0.94 |
| SVM decision value | 4.35 |
| SVM RNA-class probability | 0.999878 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2544 4785550 118 - 6264404 CUCAUGUAGCUCAGUUGGUAGAGCACACCCUUGGUAAGGGUGAGGUCAGCGGUUCAAAUCCGCUCAUGAGCUCCAUUUAUCCAGGGGCAGAUAUGAAAAUAUCUGCCCUUGUUCUA--AU ...(((.((((((.......((.(.((((((.....)))))).).))(((((.......)))))..)))))).))).....(((((((((((((....))))))))))))).....--.. ( -52.50) >pau.415 5796379 118 + 6537648 CUCAUGUAGCUCAGUUGGUAGAGCACACCCUUGGUAAGGGUGAGGUCAGCGGUUCAAAUCCGCUCAUGAGCUCCAUUUAUCCAGGGGCAGAUAUGAAAAUAUCUGCCCUUGUUCUA--AU ...(((.((((((.......((.(.((((((.....)))))).).))(((((.......)))))..)))))).))).....(((((((((((((....))))))))))))).....--.. ( -52.50) >ppk.259 5649165 118 + 6181863 CUCUUGUAGCUCAGUUGGUAGAGCACACCCUUGGUAAGGGUGAGGUCAGCGGUUCAAGUCCGCUCAAGAGCUCCAUAUAAAC-AAGGCAGAUAUGAAAAUAUCUGCCUUUGUUUUAUCA- ....((.(((((........((.(.((((((.....)))))).).))(((((.......)))))...))))).))...((((-(((((((((((....))))))))).)))))).....- ( -47.10) >pel.239 5390734 118 + 5888780 CUCUUGUAGCUCAGUUGGUAGAGCACACCCUUGGUAAGGGUGAGGUCAGCGGUUCAAAUCCGCUCAAGAGCUCCAUAUAAAC-AAGGCAGAUAUGAAAAUAUCUGCCUUUGUUUUAUCA- ....((.(((((........((.(.((((((.....)))))).).))(((((.......)))))...))))).))...((((-(((((((((((....))))))))).)))))).....- ( -47.10) >psp.2682 5244236 114 - 5928787 CUCUUGUAGCUCAGUUGGUAGAGCACACCCUUGGUAAGGGUGAGGUCAGCGGUUCAAAUCCGCUCAAGAGCUCCAUUGCUA--AAGGAAGAUAUGAAAAUAUCUGCCUUUGUUUUA---- ....((.(((((........((.(.((((((.....)))))).).))(((((.......)))))...))))).))....((--((((.((((((....)))))).)))))).....---- ( -40.50) >pst.387 5722412 114 + 6397126 CUCUUGUAGCUCAGUUGGUAGAGCACACCCUUGGUAAGGGUGAGGUCAGCGGUUCGAAUCCGCUCAAGAGCUCCAUUGCUA--AAGGCAGAUAUGAAAAUAUCUGCCUUUGUUUUA---- ....((.(((((........((.(.((((((.....)))))).).))(((((.......)))))...))))).))....((--(((((((((((....))))))))))))).....---- ( -47.10) >consensus CUCUUGUAGCUCAGUUGGUAGAGCACACCCUUGGUAAGGGUGAGGUCAGCGGUUCAAAUCCGCUCAAGAGCUCCAUUUAUAC_AAGGCAGAUAUGAAAAUAUCUGCCUUUGUUUUA__A_ ((((((..((((........)))).((((((.....)))))).....(((((.......))))))))))).............(((((((((((....)))))))))))........... (-45.09 = -44.15 + -0.94)

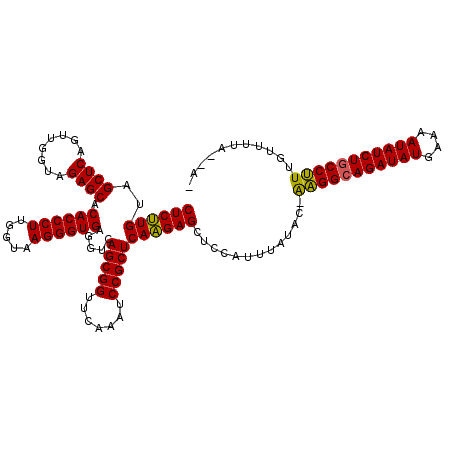
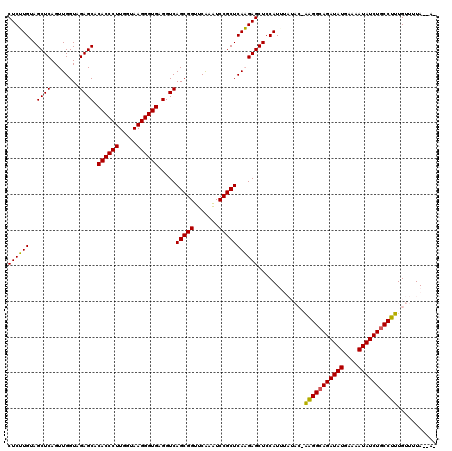
| Location | 4,785,588 – 4,785,698 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.73 |
| Mean single sequence MFE | -37.45 |
| Consensus MFE | -32.91 |
| Energy contribution | -32.47 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.900560 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2544 4785588 110 + 6264404 AUAAAUGGAGCUCAUGAGCGGAUUUGAACCGCUGACCUCACCCUUACCAAGGGUGUGCUCUACCAACUGAGCUACAUGAGCGAAA---------CACAAGCAA-CUUGACUGGAGCGGGU .........((((((((((((.......))))).....((((((.....)))))).((((........))))..)))))))....---------.((..((..-(......)..))..)) ( -35.60) >pau.415 5796417 110 - 6537648 AUAAAUGGAGCUCAUGAGCGGAUUUGAACCGCUGACCUCACCCUUACCAAGGGUGUGCUCUACCAACUGAGCUACAUGAGCGAAA---------CACAAGCAA-CUUGACUGGAGCGGGU .........((((((((((((.......))))).....((((((.....)))))).((((........))))..)))))))....---------.((..((..-(......)..))..)) ( -35.60) >ppk.259 5649203 120 - 6181863 UUAUAUGGAGCUCUUGAGCGGACUUGAACCGCUGACCUCACCCUUACCAAGGGUGUGCUCUACCAACUGAGCUACAAGAGCGAAACACUUUGUGCAAAAUCCGACAAACUUGGAGCGGGU .........((((((((((((.......))))).....((((((.....)))))).((((........))))..))))))).................(((((.((....))...))))) ( -37.10) >pss.2760 5420310 117 + 6093698 AUCAAUGGAGCUCUUGAGCGGAUUUGAACCGCUGACCUCACCCUUACCAAGGGUGUGCUCUACCAACUGAGCUACAAGAGCAAAACACUGCG--CAUGAACCA-CAAGCCUGGAGCGGGU .........((((((((((((.......))))).....((((((.....)))))).((((........))))..)))))))......((((.--......(((-......))).)))).. ( -37.10) >psp.2682 5244270 117 + 5928787 AGCAAUGGAGCUCUUGAGCGGAUUUGAACCGCUGACCUCACCCUUACCAAGGGUGUGCUCUACCAACUGAGCUACAAGAGCAAAACACAAUG--CAUGAACCA-CAAACCUGGAGCGGGU .(((.((..((((((((((((.......))))).....((((((.....)))))).((((........))))..)))))))......)).))--)........-...(((((...))))) ( -38.20) >pst.387 5722446 117 - 6397126 AGCAAUGGAGCUCUUGAGCGGAUUCGAACCGCUGACCUCACCCUUACCAAGGGUGUGCUCUACCAACUGAGCUACAAGAGCAAAACACAGUG--CAUGAACCG-CAAACAUGGAGCGGGU .(((.((..((((((((((((.......))))).....((((((.....)))))).((((........))))..)))))))......)).))--).....(((-(.........)))).. ( -41.10) >consensus AUAAAUGGAGCUCUUGAGCGGAUUUGAACCGCUGACCUCACCCUUACCAAGGGUGUGCUCUACCAACUGAGCUACAAGAGCAAAACAC__UG__CAUAAACCA_CAAACCUGGAGCGGGU .........((((((((((((.......))))).....((((((.....)))))).((((........))))..)))))))....................................... (-32.91 = -32.47 + -0.44)

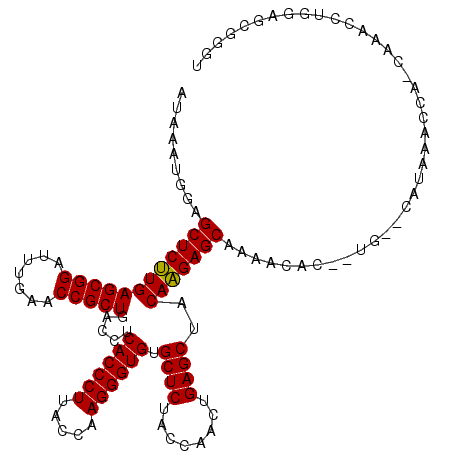
| Location | 4,785,628 – 4,785,738 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.76 |
| Mean single sequence MFE | -37.77 |
| Consensus MFE | -32.73 |
| Energy contribution | -32.73 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.602052 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2544 4785628 110 + 6264404 CCCUUACCAAGGGUGUGCUCUACCAACUGAGCUACAUGAGCGAAA---------CACAAGCAA-CUUGACUGGAGCGGGUAGCGGGAAUCGAACCCGCAUCAUCAGCUUGGAAGGCUGAG .((((.(((((.((((((((........)))).))))..((....---------.....))..-.((((.((..((((((..((.....)).))))))..)))))))))))))))..... ( -35.80) >pau.415 5796457 110 - 6537648 CCCUUACCAAGGGUGUGCUCUACCAACUGAGCUACAUGAGCGAAA---------CACAAGCAA-CUUGACUGGAGCGGGUAGCGGGAAUCGAACCCGCAUCAUCAGCUUGGAAGGCUGAG .((((.(((((.((((((((........)))).))))..((....---------.....))..-.((((.((..((((((..((.....)).))))))..)))))))))))))))..... ( -35.80) >ppk.259 5649243 120 - 6181863 CCCUUACCAAGGGUGUGCUCUACCAACUGAGCUACAAGAGCGAAACACUUUGUGCAAAAUCCGACAAACUUGGAGCGGGUAGCGGGAAUCGAACCCGCAUCAUCAGCUUGGAAGGCUGAG (((...(((((..(((((((........)))).....(.(((((....))))).)........)))..)))))...)))..(((((.......)))))....((((((.....)))))). ( -39.50) >pel.239 5390812 119 - 5888780 CCCUUACCAAGGGUGUGCUCUACCAACUGAGCUACAAGAGCGAAACACUUUGCGCAACAACUG-CAAACUUGGAGCGGGUAGCGGGAAUCGAACCCGCAUCAUCAGCUUGGAAGGCUGAG .((((.(((((((((.....))))..(((((((.((((.(((((....)))))(((.....))-)...)))).))).(((.(((((.......)))))))).)))))))))))))..... ( -42.40) >pss.2760 5420350 117 + 6093698 CCCUUACCAAGGGUGUGCUCUACCAACUGAGCUACAAGAGCAAAACACUGCG--CAUGAACCA-CAAGCCUGGAGCGGGUAGCGGGAAUCGAACCCGCAUCAUCAGCUUGGAAGGCUGAG ((((.....))))(((((((........)))).))).(.(((......))).--)........-..(((((.((((.(((.(((((.......))))))))....))))...)))))... ( -36.90) >psp.2682 5244310 117 + 5928787 CCCUUACCAAGGGUGUGCUCUACCAACUGAGCUACAAGAGCAAAACACAAUG--CAUGAACCA-CAAACCUGGAGCGGGUAGCGGGAAUCGAACCCGCAUCAUCAGCUUGGAAGGCUGAG (((...(((.(((((.((((........)))))))....(((........))--)........-....)))))...)))..(((((.......)))))....((((((.....)))))). ( -36.20) >consensus CCCUUACCAAGGGUGUGCUCUACCAACUGAGCUACAAGAGCGAAACACU_UG__CAAAAACCA_CAAACCUGGAGCGGGUAGCGGGAAUCGAACCCGCAUCAUCAGCUUGGAAGGCUGAG .((((.(((((((((.....))))..(((((((.....))).............................((..((((((..((.....)).))))))..)))))))))))))))..... (-32.73 = -32.73 + -0.00)
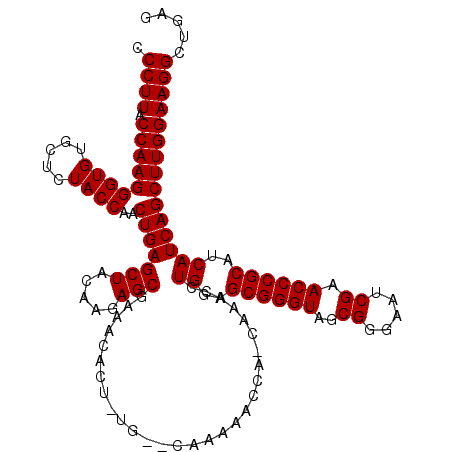
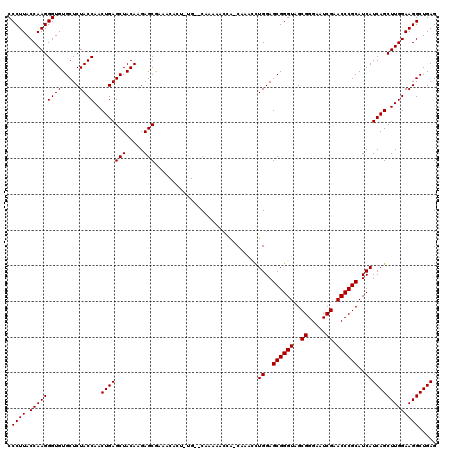
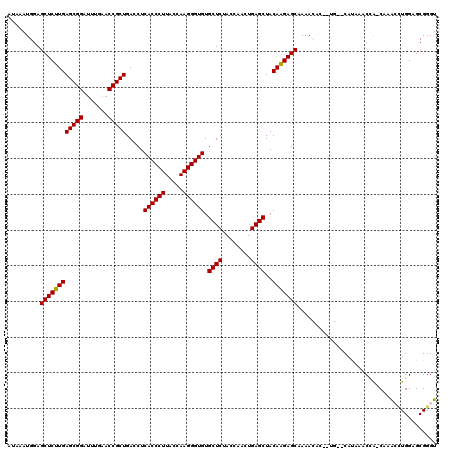
| Location | 4,785,668 – 4,785,778 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.69 |
| Mean single sequence MFE | -39.40 |
| Consensus MFE | -32.74 |
| Energy contribution | -32.88 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.867220 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2544 4785668 110 + 6264404 CGAAA---------CACAAGCAA-CUUGACUGGAGCGGGUAGCGGGAAUCGAACCCGCAUCAUCAGCUUGGAAGGCUGAGGUUCUACCACUGAACUAUACCCGCGGAGCCCGAAGCUCAC .....---------..((((...-))))......((((((((((((.......)))))....((((((.....))))))(((((.......))))).))))))).((((.....)))).. ( -40.80) >pau.415 5796497 110 - 6537648 CGAAA---------CACAAGCAA-CUUGACUGGAGCGGGUAGCGGGAAUCGAACCCGCAUCAUCAGCUUGGAAGGCUGAGGCUCUACCACUGAACUAUACCCGCGGAGCCCGAAGCUCAC .....---------..((((...-))))......((((((((((((.......)))))....((((..(((.((((....)).)).)))))))....))))))).((((.....)))).. ( -38.70) >ppk.259 5649283 120 - 6181863 CGAAACACUUUGUGCAAAAUCCGACAAACUUGGAGCGGGUAGCGGGAAUCGAACCCGCAUCAUCAGCUUGGAAGGCUGAGGUUCUACCACUAAACUAUACCCGCGGAGCUUGCGGCUCUC ...................(((((.....)))))((((((((((((.......)))))....((((((.....))))))((.....)).........)))))))(((((.....))))). ( -41.80) >pss.2760 5420390 117 + 6093698 CAAAACACUGCG--CAUGAACCA-CAAGCCUGGAGCGGGUAGCGGGAAUCGAACCCGCAUCAUCAGCUUGGAAGGCUGAGGUUCUACCACUAAACUAUACCCGCACAACUUGCAGCUCGC .......((((.--......(((-......))).((((((((((((.......)))))....((((((.....))))))((.....)).........))))))).......))))..... ( -39.50) >psp.2682 5244350 117 + 5928787 CAAAACACAAUG--CAUGAACCA-CAAACCUGGAGCGGGUAGCGGGAAUCGAACCCGCAUCAUCAGCUUGGAAGGCUGAGGUUCUACCACUAAACUAUACCCGCGCAAUUUGCAGAUCGC ..........((--((....(((-......))).((((((((((((.......)))))....((((((.....))))))((.....)).........)))))))......))))...... ( -37.80) >pst.387 5722526 117 - 6397126 CAAAACACAGUG--CAUGAACCG-CAAACAUGGAGCGGGUAGCGGGAAUCGAACCCGCAUCAUCAGCUUGGAAGGCUGAGGUUCUACCACUAAACUAUACCCGCGCAAUUUGCAGCUCGC ............--...((.(.(-((((..((..((((((((((((.......)))))....((((((.....))))))((.....)).........))))))).)).))))).).)).. ( -37.80) >consensus CAAAACAC__UG__CAUAAACCA_CAAACCUGGAGCGGGUAGCGGGAAUCGAACCCGCAUCAUCAGCUUGGAAGGCUGAGGUUCUACCACUAAACUAUACCCGCGCAACUUGCAGCUCGC ................................((((((((((((((.......)))))....((((((.....))))))((.....)).........)))))(((.....))).)))).. (-32.74 = -32.88 + 0.14)

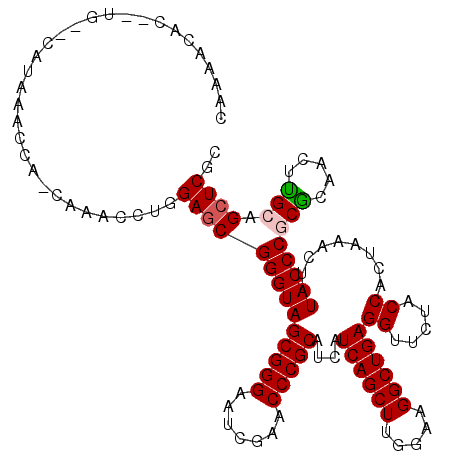
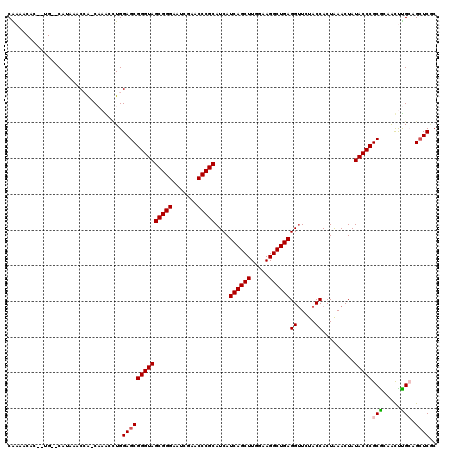
Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:10:30 2007