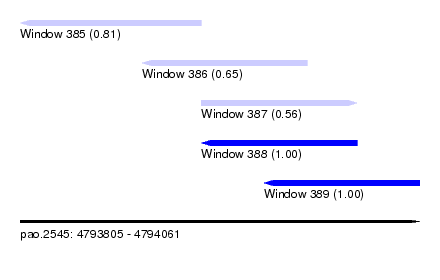
| Sequence ID | pao.2545 |
|---|---|
| Location | 4,793,805 – 4,794,061 |
| Length | 256 |
| Max. P | 0.999172 |

| Location | 4,793,805 – 4,793,921 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.16 |
| Mean single sequence MFE | -34.31 |
| Consensus MFE | -22.51 |
| Energy contribution | -22.90 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.810986 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
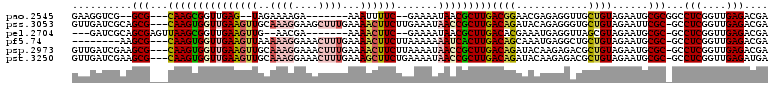
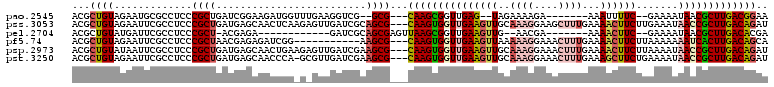
>pao.2545 4793805 116 - 6264404 AA-GC-CUUGACCAACUGCUCUUUAACAAGUCGAAUCAAGCAAUUCGUGUGGGUGCUUGUGAUGUAAGACUGGUGAUC-GCAAGAUUAUCAGCAACGCAAGUAACACU-CGUGAAUUCGA ..-((-.((((...(((...........)))....))))))((((((.(((..((((((((.((.....(((((((((-....))))))))))).)))))))).))).-..))))))... ( -33.00) >pau.414 5804636 116 + 6537648 AA-GC-CUUGACCAACUGCUCUUUAACAAGUCGAAUCAAGCAAUUCGUGUGGGUGCUUGUGAUGUAAGACUGGUGAUC-GCAAGAUUAUCAGCAGCGCAAGUAACACU-CGUGAAUUCGA ..-((-.((((...(((...........)))....))))))((((((.(((..((((((((.((.....(((((((((-....))))))))))).)))))))).))).-..))))))... ( -33.00) >pss.3053 5953248 116 - 6093698 AA-GGUCUUAACCCACCGCUCUUUAACAA-CUGAAUCAAGCAAUUCGUGUGGGUGCUUGUGGUGUCAGACUGA-AGUC-AACAGAUUAUCAGCAACCCAAGUUACUCCGCGAGAAAUCAA ..-(((........)))(((.((((....-.))))...)))..((((((.(((((((((.(((....(.((((-((((-....)))).))))).))))))).)))))))))))....... ( -29.10) >pel.2704 5003748 120 - 5888780 AACGCUCUUAACCAAUCGCUCUUUAACAAAUUGAAUCAAGCAAUUCGUGUGGGUGCUUGUGAGUACGGGCUGAUAGUCACCAAGAUUAUCAGCAUCACAAGUGGCCAUGCGAGAAAUCAC .................(((.(((((....)))))...)))..(((((((((.(((((((((......((((((((((.....)))))))))).))))))))).)))))))))....... ( -41.50) >ppk.338 5484115 120 + 6181863 AAAGCUCUUAACCAAACGCUCUUUAACAAAUCGAAUCAAGCAAUUCGUGUGGGUGCUUGUGAGUACGGACUGAUAGUCAAAAAGAUUAUCAGCAUCACAAGUGGCCAUGCGAGAAAUCAC .................(((..................)))..(((((((((.(((((((((.....(.(((((((((.....)))))))))).))))))))).)))))))))....... ( -36.97) >pf5.74 6952142 118 + 7074893 AA-GAUCUUAGCCAACCGCUCUUUAACAA-CUGAAUCAAGCAAUUCGUGUGGGUGCUUGUGGAGUCAGACUGAUAGUCAAAAAGAUUAUCAGCAUCACAAGUUACUCCGCGAGAAAUCAA ..-(((...........(((.((((....-.))))...)))..((((((.(((((((((((..(.....(((((((((.....))))))))))..)))))).)))))))))))..))).. ( -32.30) >consensus AA_GCUCUUAACCAACCGCUCUUUAACAA_UCGAAUCAAGCAAUUCGUGUGGGUGCUUGUGAUGUCAGACUGAUAGUC_AAAAGAUUAUCAGCAUCACAAGUAACACUGCGAGAAAUCAA .................(((..................)))..((((((.((.((((((((......(.(((((((((.....))))))))))..)))))))).)).))))))....... (-22.51 = -22.90 + 0.39)

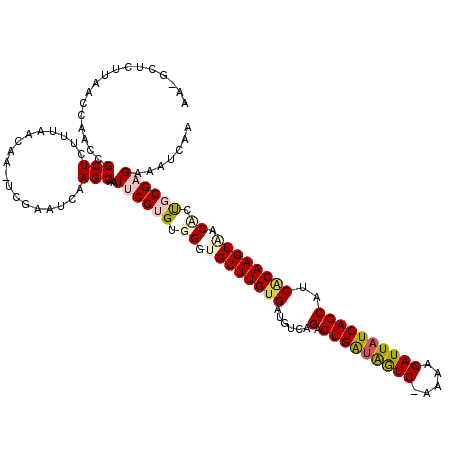
| Location | 4,793,883 – 4,793,989 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.66 |
| Mean single sequence MFE | -28.19 |
| Consensus MFE | -16.51 |
| Energy contribution | -16.48 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.652029 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4793883 106 - 6264404 ----------AAAUUUUC--GAAAAUAACGCUUGACGGAACGAGAGGUUGCUGUAGAAUGCGCGGCCUCGGUUGAGACGAAA-GC-CUUGACCAACUGCUCUUUAACAAGUCGAAUCAAG ----------.....(((--((...........(((((.....((((((((.((.....))))))))))(((..((.(....-).-))..)))..))).)).........)))))..... ( -31.75) >pss.3053 5953326 113 - 6093698 GCUUUG----AAAACUUCUUGAAAUAACCGCUUGACAGAUACAGAGGGUGCUGUAGAAUUCGC-GCCUCGGUUGAGACGAAA-GGUCUUAACCCACCGCUCUUUAACAA-CUGAAUCAAG ......----....................((((((((.((.(((((((((..........))-)))..(((((((((....-.))))))))).....)))).))....-)))..))))) ( -26.90) >pel.2704 5003828 107 - 5888780 ----------AAAACUUC--GAAAAUAACGCUUGACACGAAAUGAGGUUAGCGUAGAAUGCGC-GCCUCGGUUGAGACGAAACGCUCUUAACCAAUCGCUCUUUAACAAAUUGAAUCAAG ----------........--..........(((((..(((...(((((..((((.....))))-)))))((((((((((...)).)))))))).................)))..))))) ( -26.70) >ppk.338 5484195 117 + 6181863 UUUCUCGCAAAAAACUUC--AAAAUAAACGCUUGACAGGCUCUGAGGAAAGCGUAGAAUGCGC-GCCUCGGUUGAGACGAAAAGCUCUUAACCAAACGCUCUUUAACAAAUCGAAUCAAG ..................--.........(.((((..(((...((((...((((.....))))-.))))(((((((((.....).))))))))....)))..)))))............. ( -26.40) >pf5.74 6952222 113 + 7074893 ACUUUG----AAAACUUCUUAAAAAAAUCACUUGACAGCAAAUGAGGCUGCUGUAGAAUGCGC-GCCUCGGUUGAGACGAAA-GAUCUUAGCCAACCGCUCUUUAACAA-CUGAAUCAAG ......----....................((((((((.....((((((((........))).-)))))(((((((((....-).))))))))................-)))..))))) ( -30.80) >psp.2973 5784503 113 - 5928787 ACUUUG----AAAACUUCUUAAAAUAACCGCUUGACAGAUACAAGAGACGCUGUAGAAUGCGC-GCCUCGGUUGAGACGAAA-GCUCUUAACCACCCGCUCUUUAACAA-CUGAAUCAAG ......----....................((((((((......(((.(((.((.....))))-).)))(((((((((....-).))))))))................-)))..))))) ( -26.60) >consensus _CUUUG____AAAACUUC__AAAAAAAACGCUUGACAGAAACAGAGGAUGCUGUAGAAUGCGC_GCCUCGGUUGAGACGAAA_GCUCUUAACCAACCGCUCUUUAACAA_CUGAAUCAAG ..............................((((((((.....((((....(((.....)))...))))((((((((........)))))))).................)))..))))) (-16.51 = -16.48 + -0.02)

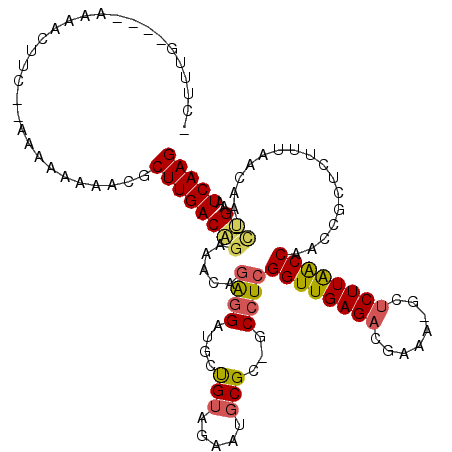
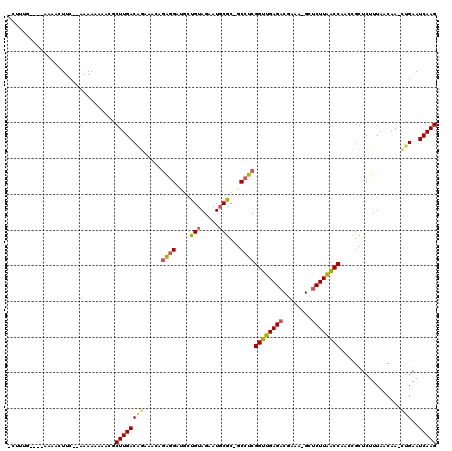
| Location | 4,793,921 – 4,794,021 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 75.53 |
| Mean single sequence MFE | -25.82 |
| Consensus MFE | -8.23 |
| Energy contribution | -8.85 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.32 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.562105 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4793921 100 + 6264404 UCGUCUCAACCGAGGCCGCGCAUUCUACAGCAACCUCUCGUUCCGUCAAGCGUUAUUUUC--GAAAAUUU-------UCUUUUCUA--CUCAACCGCUUG---CGC--CGACCUUC ..(((......((((..((..........))..))))......((.((((((........--(((((...-------..)))))..--......))))))---)).--.))).... ( -19.89) >pss.3053 5953364 112 + 6093698 UCGUCUCAACCGAGGC-GCGAAUUCUACAGCACCCUCUGUAUCUGUCAAGCGGUUAUUUCAAGAAGUUUUCAAAGCUUCCUUUGCAACUUCAACCACUUG---CGCUGCGAUCAAC (((((((....)))((-((((....(((((......)))))....))(((.((((.......((((((..(((((....))))).)))))))))).))))---))).))))..... ( -28.31) >pel.2704 5003868 101 + 5888780 UCGUCUCAACCGAGGC-GCGCAUUCUACGCUAACCUCAUUUCGUGUCAAGCGUUAUUUUC--GAAGUUUU-------UCGUU--CAACUUCAACCGCUUAACUCGCUGCGAUC--- ((((.......((((.-(((.......)))...))))....((.((.(((((........--((((((..-------.....--.))))))...))))).)).))..))))..--- ( -21.10) >pf5.74 6952260 104 - 7074893 UCGUCUCAACCGAGGC-GCGCAUUCUACAGCAGCCUCAUUUGCUGUCAAGUGAUUUUUUUAAGAAGUUUUCAAAGUUUCCUUUUUAACUUCAACCACUUG---CGCUU-------- ..(((((....)))))-((((.....(((((((......))))))).(((((.((.....))((((((...((((....))))..))))))...))))))---)))..-------- ( -29.30) >psp.2973 5784541 112 + 5928787 UCGUCUCAACCGAGGC-GCGCAUUCUACAGCGUCUCUUGUAUCUGUCAAGCGGUUAUUUUAAGAAGUUUUCAAAGUUUCCUUUGCAACUUCAACCACUUG---CGCUUCGAUCAAC ..........((((((-(((((...(((((......)))))..))).(((.((((.......((((((..(((((....))))).)))))))))).))))---)))))))...... ( -29.11) >pst.3250 6219330 112 + 6397126 UCAUCUCAACCGAGGC-GCGCAUUCUACAGCGUCUCUUGUAUCUGUCAAGCGGUUAUUUUCAGAAGCUUUCAAAGUUUCCUUUGCAACUUCAACCACUUG---CGCUUCGAUCAAC ..........((((((-(((((...(((((......)))))..))).(((.((((.......((((.((.(((((....))))).)))))))))).))))---)))))))...... ( -27.21) >consensus UCGUCUCAACCGAGGC_GCGCAUUCUACAGCAACCUCUGUAUCUGUCAAGCGGUUAUUUCAAGAAGUUUUCAAAGUUUCCUUUGCAACUUCAACCACUUG___CGCUUCGAUCAAC ..(((((....))))).(((......((((............))))................((((((.................))))))............))).......... ( -8.23 = -8.85 + 0.61)

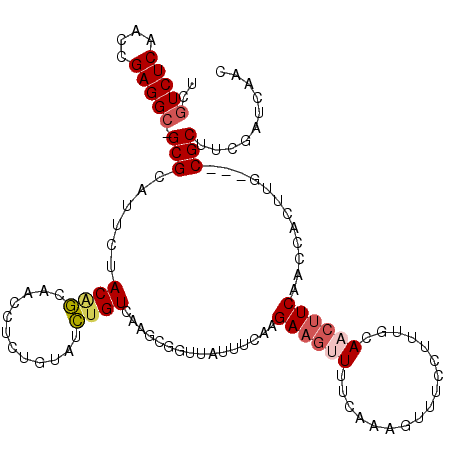
| Location | 4,793,921 – 4,794,021 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 75.53 |
| Mean single sequence MFE | -36.69 |
| Consensus MFE | -17.55 |
| Energy contribution | -18.71 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.28 |
| Mean z-score | -3.86 |
| Structure conservation index | 0.48 |
| SVM decision value | 3.20 |
| SVM RNA-class probability | 0.998711 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4793921 100 - 6264404 GAAGGUCG--GCG---CAAGCGGUUGAG--UAGAAAAGA-------AAAUUUUC--GAAAAUAACGCUUGACGGAACGAGAGGUUGCUGUAGAAUGCGCGGCCUCGGUUGAGACGA ....(((.--.((---((((((......--..(((((..-------...)))))--........)))))).)).(((..((((((((.((.....)))))))))).)))..))).. ( -32.29) >pss.3053 5953364 112 - 6093698 GUUGAUCGCAGCG---CAAGUGGUUGAAGUUGCAAAGGAAGCUUUGAAAACUUCUUGAAAUAACCGCUUGACAGAUACAGAGGGUGCUGUAGAAUUCGC-GCCUCGGUUGAGACGA .(..((((..(((---(((((((((((((((.(((((....)))))..)))))).......))))))))((....(((((......)))))....))))-))..))))..)..... ( -37.31) >pel.2704 5003868 101 - 5888780 ---GAUCGCAGCGAGUUAAGCGGUUGAAGUUG--AACGA-------AAAACUUC--GAAAAUAACGCUUGACACGAAAUGAGGUUAGCGUAGAAUGCGC-GCCUCGGUUGAGACGA ---..((.((((..((((((((.((((((((.--.....-------..))))))--))......))))))))......((((((..((((.....))))-)))))))))).))... ( -36.30) >pf5.74 6952260 104 + 7074893 --------AAGCG---CAAGUGGUUGAAGUUAAAAAGGAAACUUUGAAAACUUCUUAAAAAAAUCACUUGACAGCAAAUGAGGCUGCUGUAGAAUGCGC-GCCUCGGUUGAGACGA --------..((.---(((((((((((((((..((((....))))...)))))).......)))))))))...))...(((((((((........))).-)))))).......... ( -35.21) >psp.2973 5784541 112 - 5928787 GUUGAUCGAAGCG---CAAGUGGUUGAAGUUGCAAAGGAAACUUUGAAAACUUCUUAAAAUAACCGCUUGACAGAUACAAGAGACGCUGUAGAAUGCGC-GCCUCGGUUGAGACGA .(..(((((.(((---(((((((((((((((.(((((....)))))..)))))).......))))))))(.((..((((........))))...)))))-)).)))))..)..... ( -39.51) >pst.3250 6219330 112 - 6397126 GUUGAUCGAAGCG---CAAGUGGUUGAAGUUGCAAAGGAAACUUUGAAAGCUUCUGAAAAUAACCGCUUGACAGAUACAAGAGACGCUGUAGAAUGCGC-GCCUCGGUUGAGAUGA .(..(((((.(((---(((((((((((((((.(((((....)))))..)))))).......))))))))(.((..((((........))))...)))))-)).)))))..)..... ( -39.51) >consensus GUUGAUCGAAGCG___CAAGUGGUUGAAGUUGCAAAGGAAACUUUGAAAACUUCUUAAAAAAACCGCUUGACAGAAACAGAGGAUGCUGUAGAAUGCGC_GCCUCGGUUGAGACGA ..........(((...(((((((((((((((..((((....))))...)))))).......)))))))))((((............))))......)))...(((....))).... (-17.55 = -18.71 + 1.16)
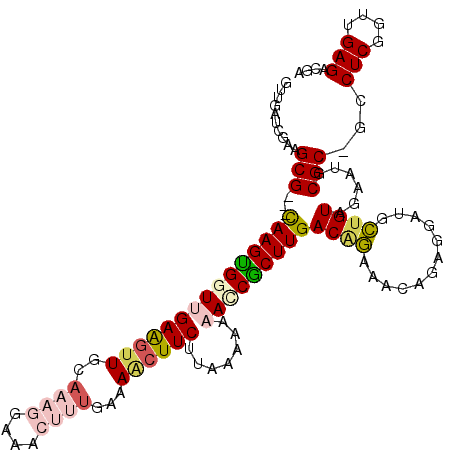
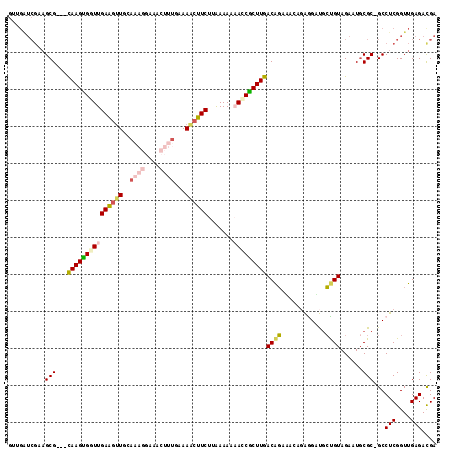
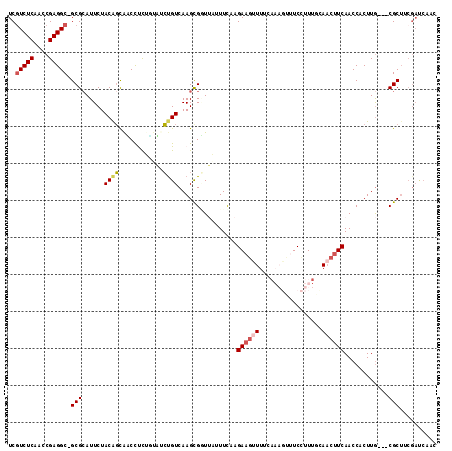
| Location | 4,793,961 – 4,794,061 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 72.47 |
| Mean single sequence MFE | -35.09 |
| Consensus MFE | -16.71 |
| Energy contribution | -18.29 |
| Covariance contribution | 1.57 |
| Combinations/Pair | 1.26 |
| Mean z-score | -3.38 |
| Structure conservation index | 0.48 |
| SVM decision value | 3.41 |
| SVM RNA-class probability | 0.999172 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4793961 100 - 6264404 ACGCUGUAGAAUGCGCCUCCCGCUGAUCGGAAGAUGGUUUGAAGGUCG--GCG---CAAGCGGUUGAG--UAGAAAAGA-------AAAUUUUC--GAAAAUAACGCUUGACGGAA ...((((.....(((((..(((((.(((....)))))).....))..)--)))---)(((((......--..(((((..-------...)))))--........))))).)))).. ( -29.79) >pss.3053 5953403 113 - 6093698 ACGCUGUAGAAUUCGCCUCCCGCUGAUGAGCAACUCAAGAGUUGAUCGCAGCG---CAAGUGGUUGAAGUUGCAAAGGAAGCUUUGAAAACUUCUUGAAAUAACCGCUUGACAGAU ...((((.............(((((.(((.(((((....))))).))))))))---(((((((((((((((.(((((....)))))..)))))).......))))))))))))).. ( -39.61) >pel.2704 5003907 92 - 5888780 ACGCUGUAUGAUUCGCCUCCCGCU-ACGAGA------------GAUCGCAGCGAGUUAAGCGGUUGAAGUUG--AACGA-------AAAACUUC--GAAAAUAACGCUUGACACGA .((((((..(((.(..(((.....-..))).------------)))))))))).((((((((.((((((((.--.....-------..))))))--))......)))))))).... ( -30.80) >pf5.74 6952299 102 + 7074893 ACGCUGUAGAAUUCGCCUCCCGCUAACGAGAGAUCGG-----------AAGCG---CAAGUGGUUGAAGUUAAAAAGGAAACUUUGAAAACUUCUUAAAAAAAUCACUUGACAGCA ..(((((.............((((..(((....))).-----------.))))---(((((((((((((((..((((....))))...)))))).......)))))))))))))). ( -35.61) >psp.2973 5784580 113 - 5928787 ACGCUGUAUAAUUCGCCUCCCGCUGAUGAGCAACUGAAGAGUUGAUCGAAGCG---CAAGUGGUUGAAGUUGCAAAGGAAACUUUGAAAACUUCUUAAAAUAACCGCUUGACAGAU ...((((.............((((..(((.(((((....))))).))).))))---(((((((((((((((.(((((....)))))..)))))).......))))))))))))).. ( -38.31) >pst.3250 6219369 112 - 6397126 ACGCUGUAGAAUUCGCCUCCCGCUGAUGAGCAACCCA-GCGUUGAUCGAAGCG---CAAGUGGUUGAAGUUGCAAAGGAAACUUUGAAAGCUUCUGAAAAUAACCGCUUGACAGAU ...((((.............((((..(((.((((...-..)))).))).))))---(((((((((((((((.(((((....)))))..)))))).......))))))))))))).. ( -36.41) >consensus ACGCUGUAGAAUUCGCCUCCCGCUGAUGAGAAACUGA_G_GUUGAUCGAAGCG___CAAGUGGUUGAAGUUGCAAAGGAAACUUUGAAAACUUCUUAAAAAAACCGCUUGACAGAA ...((((.............((((.........................))))...(((((((((((((((..((((....))))...)))))).......))))))))))))).. (-16.71 = -18.29 + 1.57)
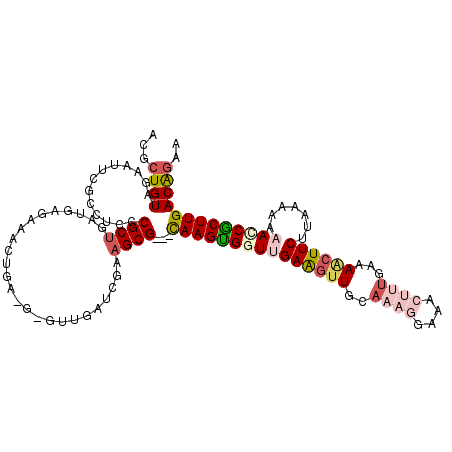
Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:10:24 2007