
| Sequence ID | pao.2545 |
|---|---|
| Location | 4,792,821 – 4,793,660 |
| Length | 839 |
| Max. P | 0.999996 |

| Location | 4,792,821 – 4,792,941 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.44 |
| Mean single sequence MFE | -43.30 |
| Consensus MFE | -44.63 |
| Energy contribution | -43.30 |
| Covariance contribution | -1.33 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.99 |
| Structure conservation index | 1.03 |
| SVM decision value | 4.75 |
| SVM RNA-class probability | 0.999946 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
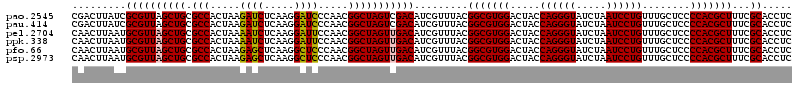
>pao.2545 4792821 120 - 6264404 UGGUAGUCCACGCCGUAAACGAUGUCGACUAGCCGUUGGGAUCCUUGAGAUCUUAGUGGCGCAGCUAACGCGAUAAGUCGACCGCCUGGGGAGUACGGCCGCAAGGUUAAAACUCAAAUG ......((((.((((....))..(((((((.((((((((((((.....))))))))))))...((....))....))))))).)).))))((((..((((....))))...))))..... ( -44.60) >pau.414 5803652 120 + 6537648 UGGUAGUCCACGCCGUAAACGAUGUCGACUAGCCGUUGGGAUCCUUGAGAUCUUAGUGGCGCAGCUAACGCGAUAAGUCGACCGCCUGGGGAGUACGGCCGCAAGGUUAAAACUCAAAUG ......((((.((((....))..(((((((.((((((((((((.....))))))))))))...((....))....))))))).)).))))((((..((((....))))...))))..... ( -44.60) >pel.2704 5002779 120 - 5888780 UGGUAGUCCACGCCGUAAACGAUGUCAACUAGCCGUUGGAAUCCUUGAGAUUUUAGUGGCGCAGCUAACGCAUUAAGUUGACCGCCUGGGGAGUACGGCCGCAAGGUUAAAACUCAAAUG ......((((.((((....))..(((((((.((((((((((((.....))))))))))))...((....))....))))))).)).))))((((..((((....))))...))))..... ( -40.50) >ppk.338 5483146 120 + 6181863 UGGUAGUCCACGCCGUAAACGAUGUCAACUAGCCGUUGGAAUCCUUGAGAUUUUAGUGGCGCAGCUAACGCAUUAAGUUGACCGCCUGGGGAGUACGGCCGCAAGGUUAAAACUCAAAUG ......((((.((((....))..(((((((.((((((((((((.....))))))))))))...((....))....))))))).)).))))((((..((((....))))...))))..... ( -40.50) >pfo.66 6316028 120 + 6438405 UGGUAGUCCACGCCGUAAACGAUGUCAACUAGCCGUUGGGAGCCUUGAGCUCUUAGUGGCGCAGCUAACGCAUUAAGUUGACCGCCUGGGGAGUACGGCCGCAAGGUUAAAACUCAAAUG ......((((.((((....))..(((((((.((((((((((((.....))))))))))))...((....))....))))))).)).))))((((..((((....))))...))))..... ( -44.80) >psp.2973 5783451 120 - 5928787 UGGUAGUCCACGCCGUAAACGAUGUCAACUAGCCGUUGGGAGCCUUGAGCUCUUAGUGGCGCAGCUAACGCAUUAAGUUGACCGCCUGGGGAGUACGGCCGCAAGGUUAAAACUCAAAUG ......((((.((((....))..(((((((.((((((((((((.....))))))))))))...((....))....))))))).)).))))((((..((((....))))...))))..... ( -44.80) >consensus UGGUAGUCCACGCCGUAAACGAUGUCAACUAGCCGUUGGGAUCCUUGAGAUCUUAGUGGCGCAGCUAACGCAUUAAGUUGACCGCCUGGGGAGUACGGCCGCAAGGUUAAAACUCAAAUG ......((((.((((....))..(((((((.((((((((((((.....))))))))))))...((....))....))))))).)).))))((((..((((....))))...))))..... (-44.63 = -43.30 + -1.33)


| Location | 4,792,861 – 4,792,981 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.44 |
| Mean single sequence MFE | -35.92 |
| Consensus MFE | -36.27 |
| Energy contribution | -35.72 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.98 |
| Structure conservation index | 1.01 |
| SVM decision value | 4.40 |
| SVM RNA-class probability | 0.999889 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4792861 120 + 6264404 CGACUUAUCGCGUUAGCUGCGCCACUAAGAUCUCAAGGAUCCCAACGGCUAGUCGACAUCGUUUACGGCGUGGACUACCAGGGUAUCUAAUCCUGUUUGCUCCCCACGCUUUCGCACCUC .........((((..((((.(((.....((((.....)))).....)))))))..)).........(((((((.....((((((.....))))))........)))))))...))..... ( -34.12) >pau.414 5803692 120 - 6537648 CGACUUAUCGCGUUAGCUGCGCCACUAAGAUCUCAAGGAUCCCAACGGCUAGUCGACAUCGUUUACGGCGUGGACUACCAGGGUAUCUAAUCCUGUUUGCUCCCCACGCUUUCGCACCUC .........((((..((((.(((.....((((.....)))).....)))))))..)).........(((((((.....((((((.....))))))........)))))))...))..... ( -34.12) >pel.2704 5002819 120 + 5888780 CAACUUAAUGCGUUAGCUGCGCCACUAAAAUCUCAAGGAUUCCAACGGCUAGUUGACAUCGUUUACGGCGUGGACUACCAGGGUAUCUAAUCCUGUUUGCUCCCCACGCUUUCGCACCUC ........(((((((((((.(((.....((((.....)))).....))))))))))).........(((((((.....((((((.....))))))........)))))))...))).... ( -34.72) >ppk.338 5483186 120 - 6181863 CAACUUAAUGCGUUAGCUGCGCCACUAAAAUCUCAAGGAUUCCAACGGCUAGUUGACAUCGUUUACGGCGUGGACUACCAGGGUAUCUAAUCCUGUUUGCUCCCCACGCUUUCGCACCUC ........(((((((((((.(((.....((((.....)))).....))))))))))).........(((((((.....((((((.....))))))........)))))))...))).... ( -34.72) >pfo.66 6316068 120 - 6438405 CAACUUAAUGCGUUAGCUGCGCCACUAAGAGCUCAAGGCUCCCAACGGCUAGUUGACAUCGUUUACGGCGUGGACUACCAGGGUAUCUAAUCCUGUUUGCUCCCCACGCUUUCGCACCUC ........(((((((((((.(((.....((((.....)))).....))))))))))).........(((((((.....((((((.....))))))........)))))))...))).... ( -38.92) >psp.2973 5783491 120 + 5928787 CAACUUAAUGCGUUAGCUGCGCCACUAAGAGCUCAAGGCUCCCAACGGCUAGUUGACAUCGUUUACGGCGUGGACUACCAGGGUAUCUAAUCCUGUUUGCUCCCCACGCUUUCGCACCUC ........(((((((((((.(((.....((((.....)))).....))))))))))).........(((((((.....((((((.....))))))........)))))))...))).... ( -38.92) >consensus CAACUUAAUGCGUUAGCUGCGCCACUAAGAUCUCAAGGAUCCCAACGGCUAGUUGACAUCGUUUACGGCGUGGACUACCAGGGUAUCUAAUCCUGUUUGCUCCCCACGCUUUCGCACCUC .........((((((((((.(((.....((((.....)))).....))))))))))).........(((((((.....((((((.....))))))........)))))))...))..... (-36.27 = -35.72 + -0.55)
| Location | 4,792,861 – 4,792,981 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.44 |
| Mean single sequence MFE | -40.30 |
| Consensus MFE | -41.63 |
| Energy contribution | -40.30 |
| Covariance contribution | -1.33 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.30 |
| Structure conservation index | 1.03 |
| SVM decision value | 2.86 |
| SVM RNA-class probability | 0.997431 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
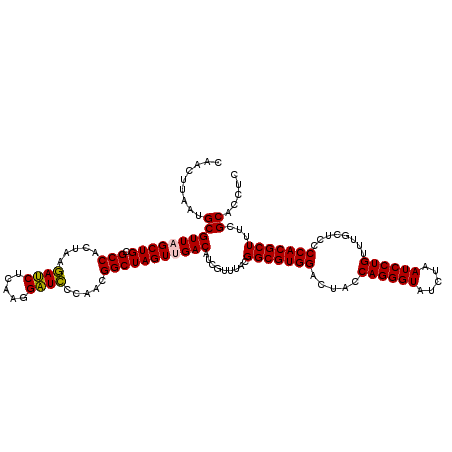
>pao.2545 4792861 120 - 6264404 GAGGUGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGUCGACUAGCCGUUGGGAUCCUUGAGAUCUUAGUGGCGCAGCUAACGCGAUAAGUCG ..(((((....))(((((..((...((((.........))))))..))))))))...........(((((.((((((((((((.....))))))))))))...((....))....))))) ( -41.60) >pau.414 5803692 120 + 6537648 GAGGUGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGUCGACUAGCCGUUGGGAUCCUUGAGAUCUUAGUGGCGCAGCUAACGCGAUAAGUCG ..(((((....))(((((..((...((((.........))))))..))))))))...........(((((.((((((((((((.....))))))))))))...((....))....))))) ( -41.60) >pel.2704 5002819 120 - 5888780 GAGGUGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGUCAACUAGCCGUUGGAAUCCUUGAGAUUUUAGUGGCGCAGCUAACGCAUUAAGUUG ..(((((....))(((((..((...((((.........))))))..))))))))...........(((((.((((((((((((.....))))))))))))...((....))....))))) ( -37.50) >ppk.338 5483186 120 + 6181863 GAGGUGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGUCAACUAGCCGUUGGAAUCCUUGAGAUUUUAGUGGCGCAGCUAACGCAUUAAGUUG ..(((((....))(((((..((...((((.........))))))..))))))))...........(((((.((((((((((((.....))))))))))))...((....))....))))) ( -37.50) >pfo.66 6316068 120 + 6438405 GAGGUGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGUCAACUAGCCGUUGGGAGCCUUGAGCUCUUAGUGGCGCAGCUAACGCAUUAAGUUG ..(((((....))(((((..((...((((.........))))))..))))))))...........(((((.((((((((((((.....))))))))))))...((....))....))))) ( -41.80) >psp.2973 5783491 120 - 5928787 GAGGUGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGUCAACUAGCCGUUGGGAGCCUUGAGCUCUUAGUGGCGCAGCUAACGCAUUAAGUUG ..(((((....))(((((..((...((((.........))))))..))))))))...........(((((.((((((((((((.....))))))))))))...((....))....))))) ( -41.80) >consensus GAGGUGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGUCAACUAGCCGUUGGGAUCCUUGAGAUCUUAGUGGCGCAGCUAACGCAUUAAGUUG ..(((((....))(((((..((...((((.........))))))..))))))))...........(((((.((((((((((((.....))))))))))))...((....))....))))) (-41.63 = -40.30 + -1.33)

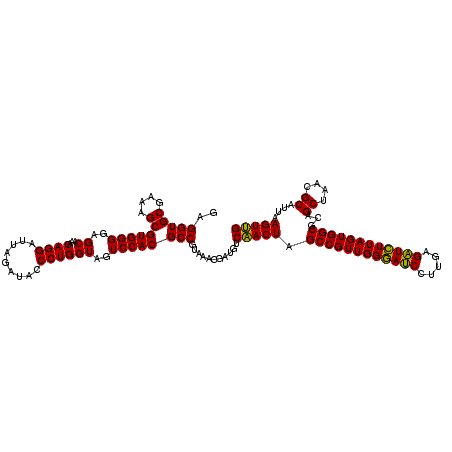
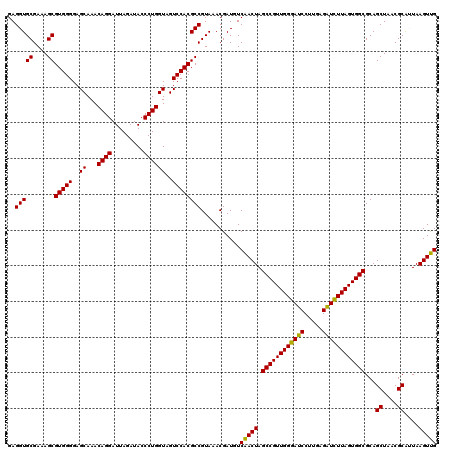
| Location | 4,792,941 – 4,793,061 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -35.90 |
| Consensus MFE | -35.90 |
| Energy contribution | -35.90 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.94 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.894751 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4792941 120 - 6264404 UUCCUGUGUAGCGGUGAAAUGCGUAGAUAUAGGAAGGAACACCAGUGGCGAAGGCGACCACCUGGACUGAUACUGACACUGAGGUGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCC (((((((((.(((......)))....)))))))))((....(((((((((....)).))).)))).......((((..(((....((....))((.....))...)))..))))...)). ( -35.90) >pel.2704 5002899 120 - 5888780 UUCCUGUGUAGCGGUGAAAUGCGUAGAUAUAGGAAGGAACACCAGUGGCGAAGGCGACCACCUGGACUGAUACUGACACUGAGGUGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCC (((((((((.(((......)))....)))))))))((....(((((((((....)).))).)))).......((((..(((....((....))((.....))...)))..))))...)). ( -35.90) >ppk.338 5483266 120 + 6181863 UUCCUGUGUAGCGGUGAAAUGCGUAGAUAUAGGAAGGAACACCAGUGGCGAAGGCGACCACCUGGACUGAUACUGACACUGAGGUGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCC (((((((((.(((......)))....)))))))))((....(((((((((....)).))).)))).......((((..(((....((....))((.....))...)))..))))...)). ( -35.90) >pf5.74 6951290 120 + 7074893 UUCCUGUGUAGCGGUGAAAUGCGUAGAUAUAGGAAGGAACACCAGUGGCGAAGGCGACCACCUGGACUGAUACUGACACUGAGGUGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCC (((((((((.(((......)))....)))))))))((....(((((((((....)).))).)))).......((((..(((....((....))((.....))...)))..))))...)). ( -35.90) >pfo.66 6316148 120 + 6438405 UUCCUGUGUAGCGGUGAAAUGCGUAGAUAUAGGAAGGAACACCAGUGGCGAAGGCGACCACCUGGACUGAUACUGACACUGAGGUGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCC (((((((((.(((......)))....)))))))))((....(((((((((....)).))).)))).......((((..(((....((....))((.....))...)))..))))...)). ( -35.90) >psp.2973 5783571 120 - 5928787 UUCCUGUGUAGCGGUGAAAUGCGUAGAUAUAGGAAGGAACACCAGUGGCGAAGGCGACCACCUGGACUGAUACUGACACUGAGGUGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCC (((((((((.(((......)))....)))))))))((....(((((((((....)).))).)))).......((((..(((....((....))((.....))...)))..))))...)). ( -35.90) >consensus UUCCUGUGUAGCGGUGAAAUGCGUAGAUAUAGGAAGGAACACCAGUGGCGAAGGCGACCACCUGGACUGAUACUGACACUGAGGUGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCC (((((((((.(((......)))....)))))))))((....(((((((((....)).))).)))).......((((..(((....((....))((.....))...)))..))))...)). (-35.90 = -35.90 + -0.00)

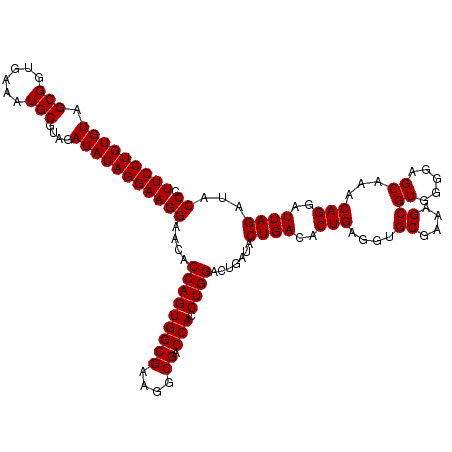
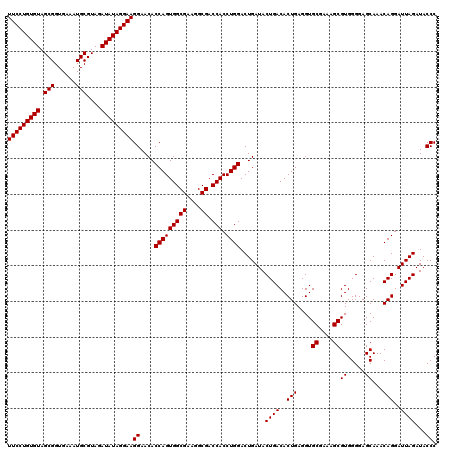
| Location | 4,792,981 – 4,793,101 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.72 |
| Mean single sequence MFE | -41.85 |
| Consensus MFE | -41.29 |
| Energy contribution | -41.15 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.12 |
| Structure conservation index | 0.99 |
| SVM decision value | 5.52 |
| SVM RNA-class probability | 0.999989 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4792981 120 - 6264404 CAAAACUACUGAGCUAGAGUACGGUAGAGGGUGGUGGAAUUUCCUGUGUAGCGGUGAAAUGCGUAGAUAUAGGAAGGAACACCAGUGGCGAAGGCGACCACCUGGACUGAUACUGACACU .................((((((((..(((.(((((...((((((((((.(((......)))....))))))))))...)))))((.((....)).))..)))..)))).))))...... ( -40.20) >pau.414 5803812 120 + 6537648 CAAAACUACUGAGCUAGAGUACGGUAGAGGGUGGUGGAAUUUCCUGUGUAGCGGUGAAAUGCGUAGAUAUAGGAAGGAACACCAGUGGCGAAGGCGACCACCUGGACUGAUACUGACACU .................((((((((..(((.(((((...((((((((((.(((......)))....))))))))))...)))))((.((....)).))..)))..)))).))))...... ( -40.20) >pel.2704 5002939 120 - 5888780 CAAAACUGGCGAGCUAGAGUAUGGUAGAGGGUGGUGGAAUUUCCUGUGUAGCGGUGAAAUGCGUAGAUAUAGGAAGGAACACCAGUGGCGAAGGCGACCACCUGGACUGAUACUGACACU .....((((....))))((((((((..(((.(((((...((((((((((.(((......)))....))))))))))...)))))((.((....)).))..)))..))).)))))...... ( -42.30) >ppk.338 5483306 120 + 6181863 CAAAACUGGCAAGCUAGAGUACGGUAGAGGGUGGUGGAAUUUCCUGUGUAGCGGUGAAAUGCGUAGAUAUAGGAAGGAACACCAGUGGCGAAGGCGACCACCUGGACUGAUACUGACACU .....((((....))))((((((((..(((.(((((...((((((((((.(((......)))....))))))))))...)))))((.((....)).))..)))..)))).))))...... ( -43.80) >pfo.66 6316188 120 + 6438405 CAAAACUGGCGAGCUAGAGUAUGGUAGAGGGUGGUGGAAUUUCCUGUGUAGCGGUGAAAUGCGUAGAUAUAGGAAGGAACACCAGUGGCGAAGGCGACCACCUGGACUGAUACUGACACU .....((((....))))((((((((..(((.(((((...((((((((((.(((......)))....))))))))))...)))))((.((....)).))..)))..))).)))))...... ( -42.30) >psp.2973 5783611 120 - 5928787 CAAAACUGGCAAGCUAGAGUAUGGUAGAGGGUGGUGGAAUUUCCUGUGUAGCGGUGAAAUGCGUAGAUAUAGGAAGGAACACCAGUGGCGAAGGCGACCACCUGGACUGAUACUGACACU .....((((....))))((((((((..(((.(((((...((((((((((.(((......)))....))))))))))...)))))((.((....)).))..)))..))).)))))...... ( -42.30) >consensus CAAAACUGGCGAGCUAGAGUACGGUAGAGGGUGGUGGAAUUUCCUGUGUAGCGGUGAAAUGCGUAGAUAUAGGAAGGAACACCAGUGGCGAAGGCGACCACCUGGACUGAUACUGACACU .....((((....))))((((((((..(((.(((((...((((((((((.(((......)))....))))))))))...)))))((.((....)).))..)))..)))).))))...... (-41.29 = -41.15 + -0.14)

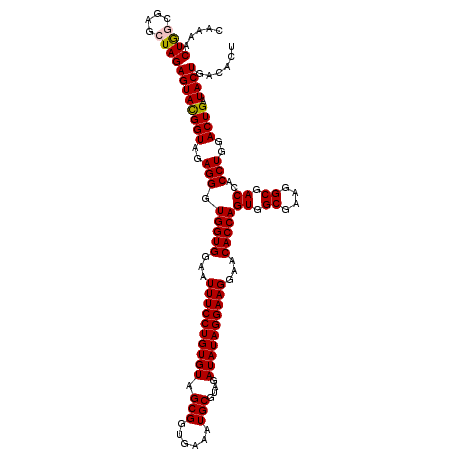
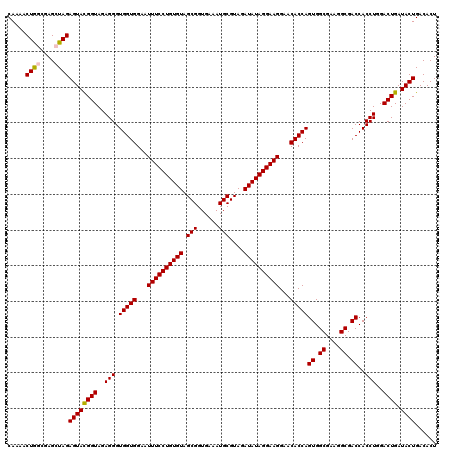
| Location | 4,793,021 – 4,793,141 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.89 |
| Mean single sequence MFE | -37.72 |
| Consensus MFE | -35.49 |
| Energy contribution | -35.05 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.62 |
| SVM RNA-class probability | 0.995819 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4793021 120 - 6264404 UUGGAUGUGAAAUCCCCGGGCUCAACCUGGGAACUGCAUCCAAAACUACUGAGCUAGAGUACGGUAGAGGGUGGUGGAAUUUCCUGUGUAGCGGUGAAAUGCGUAGAUAUAGGAAGGAAC (((((((((...((((.((......)).))))..)))))))))..((((((.((....)).))))))............((((((((((.(((......)))....)))))))))).... ( -41.00) >pau.414 5803852 120 + 6537648 UUGGAUGUGAAAUCCCCGGGCUCAACCUGGGAACUGCAUCCAAAACUACUGAGCUAGAGUACGGUAGAGGGUGGUGGAAUUUCCUGUGUAGCGGUGAAAUGCGUAGAUAUAGGAAGGAAC (((((((((...((((.((......)).))))..)))))))))..((((((.((....)).))))))............((((((((((.(((......)))....)))))))))).... ( -41.00) >pss.3053 5952474 120 - 6093698 UUGAAUGUGAAAUCCCCGGGCUCAACCUGGGAACUGCAUCCAAAACUGGCAAGCUAGAGUAUGGUAGAGGGUGGUGGAAUUUCCUGUGUAGCGGUGAAAUGCGUAGAUAUAGGAAGGAAC ............((((((..(((.(((..(((......)))....((((....)))).....))).)))..))).))).((((((((((.(((......)))....)))))))))).... ( -34.00) >pel.2704 5002979 120 - 5888780 UUGGAUGUGAAAGCCCCGGGCUCAACCUGGGAACUGCAUCCAAAACUGGCGAGCUAGAGUAUGGUAGAGGGUGGUGGAAUUUCCUGUGUAGCGGUGAAAUGCGUAGAUAUAGGAAGGAAC ((((((((......((((((.....))))))....))))))))..(((.((.((....)).)).)))............((((((((((.(((......)))....)))))))))).... ( -36.60) >ppk.338 5483346 120 + 6181863 UUGGAUGUGAAAGCCCCGGGCUCAACCUGGGAACUGCAUCCAAAACUGGCAAGCUAGAGUACGGUAGAGGGUGGUGGAAUUUCCUGUGUAGCGGUGAAAUGCGUAGAUAUAGGAAGGAAC ((((((((......((((((.....))))))....))))))))..((((....))))(.(((........))).)....((((((((((.(((......)))....)))))))))).... ( -36.40) >pf5.74 6951370 120 + 7074893 UUGGAUGUGAAAGCCCCGGGCUCAACCUGGGAACUGCAUCCAAAACUGGCAAGCUAGAGUAUGGUAGAGGGUGGUGGAAUUUCCUGUGUAGCGGUGAAAUGCGUAGAUAUAGGAAGGAAC ((((((((......((((((.....))))))....))))))))..(((.((.((....)).)).)))............((((((((((.(((......)))....)))))))))).... ( -37.30) >consensus UUGGAUGUGAAAGCCCCGGGCUCAACCUGGGAACUGCAUCCAAAACUGGCAAGCUAGAGUACGGUAGAGGGUGGUGGAAUUUCCUGUGUAGCGGUGAAAUGCGUAGAUAUAGGAAGGAAC ((((((((......((((((.....))))))....))))))))..(((.((.((....)).)).)))............((((((((((.(((......)))....)))))))))).... (-35.49 = -35.05 + -0.44)

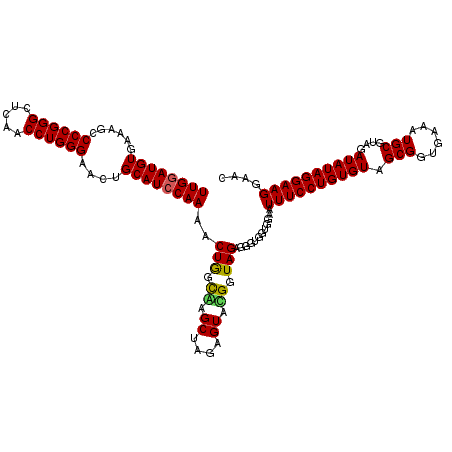
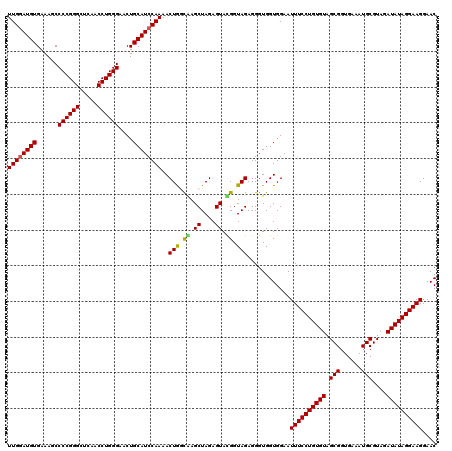
| Location | 4,793,061 – 4,793,181 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.06 |
| Mean single sequence MFE | -39.75 |
| Consensus MFE | -40.65 |
| Energy contribution | -39.52 |
| Covariance contribution | -1.14 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.71 |
| Structure conservation index | 1.02 |
| SVM decision value | 3.93 |
| SVM RNA-class probability | 0.999711 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
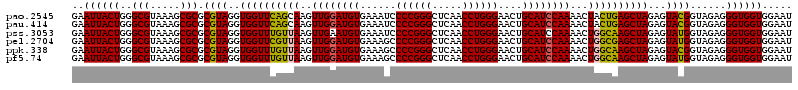
>pao.2545 4793061 120 - 6264404 GAAUUACUGGGCGUAAAGCGCGCGUAGGUGGUUCAGCAAGUUGGAUGUGAAAUCCCCGGGCUCAACCUGGGAACUGCAUCCAAAACUACUGAGCUAGAGUACGGUAGAGGGUGGUGGAAU (((((((((.((((.....)))).)).))))))).((...(((((((((...((((.((......)).))))..)))))))))..((((((.((....)).))))))...))........ ( -41.50) >pau.414 5803892 120 + 6537648 GAAUUACUGGGCGUAAAGCGCGCGUAGGUGGUUCAGCAAGUUGGAUGUGAAAUCCCCGGGCUCAACCUGGGAACUGCAUCCAAAACUACUGAGCUAGAGUACGGUAGAGGGUGGUGGAAU (((((((((.((((.....)))).)).))))))).((...(((((((((...((((.((......)).))))..)))))))))..((((((.((....)).))))))...))........ ( -41.50) >pss.3053 5952514 120 - 6093698 GAAUUACUGGGCGUAAAGCGCGCGUAGGUGGUUUGUUAAGUUGAAUGUGAAAUCCCCGGGCUCAACCUGGGAACUGCAUCCAAAACUGGCAAGCUAGAGUAUGGUAGAGGGUGGUGGAAU ..(((((((.(((.....))).).....((((((((((..(((.(((((...((((.((......)).))))..))))).)))...)))))))))).............))))))..... ( -33.10) >pel.2704 5003019 120 - 5888780 GAAUUACUGGGCGUAAAGCGCGCGUAGGUGGUUCGUUAAGUUGGAUGUGAAAGCCCCGGGCUCAACCUGGGAACUGCAUCCAAAACUGGCGAGCUAGAGUAUGGUAGAGGGUGGUGGAAU ..(((((((.(((.....))).).....((((((((((..((((((((......((((((.....))))))....))))))))...)))))))))).............))))))..... ( -41.50) >ppk.338 5483386 120 + 6181863 GAAUUACUGGGCGUAAAGCGCGCGUAGGUGGUUUGUUAAGUUGGAUGUGAAAGCCCCGGGCUCAACCUGGGAACUGCAUCCAAAACUGGCAAGCUAGAGUACGGUAGAGGGUGGUGGAAU ..((((((..(((.....))).((((..((((((((((..((((((((......((((((.....))))))....))))))))...))))))))))...))))......))))))..... ( -41.20) >pf5.74 6951410 120 + 7074893 GAAUUACUGGGCGUAAAGCGCGCGUAGGUGGUUUGUUAAGUUGGAUGUGAAAGCCCCGGGCUCAACCUGGGAACUGCAUCCAAAACUGGCAAGCUAGAGUAUGGUAGAGGGUGGUGGAAU ..(((((((.(((.....))).).....((((((((((..((((((((......((((((.....))))))....))))))))...)))))))))).............))))))..... ( -39.70) >consensus GAAUUACUGGGCGUAAAGCGCGCGUAGGUGGUUCGUUAAGUUGGAUGUGAAAGCCCCGGGCUCAACCUGGGAACUGCAUCCAAAACUGGCAAGCUAGAGUACGGUAGAGGGUGGUGGAAU ..((((((..(((.....))).((((..((((((((((..((((((((......((((((.....))))))....))))))))...))))))))))...))))......))))))..... (-40.65 = -39.52 + -1.14)
| Location | 4,793,181 – 4,793,301 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.39 |
| Mean single sequence MFE | -37.45 |
| Consensus MFE | -41.00 |
| Energy contribution | -37.45 |
| Covariance contribution | -3.55 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.36 |
| Structure conservation index | 1.09 |
| SVM decision value | 3.51 |
| SVM RNA-class probability | 0.999315 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
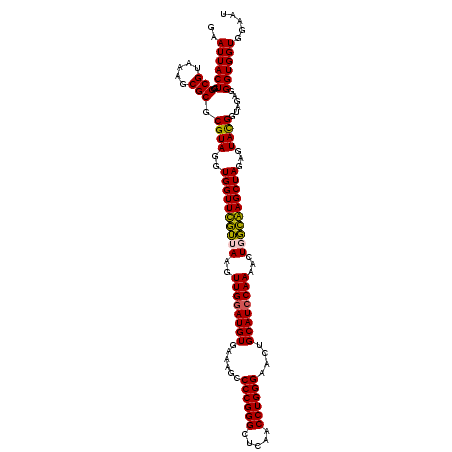
>pao.2545 4793181 120 - 6264404 UUAAGUUGGGAGGAAGGGCAGUAAGUUAAUACCUUGCUGUUUUGACGUUACCAACAGAAUAAGCACCGGCUAACUUCGUGCCAGCAGCCGCGGUAAUACGAAGGGUGCAAGCGUUAAUCG ....(((((..(..(((((((((((.......)))))))))))..)....))))).......(((((......(((((((((.((....)))))...)))))))))))............ ( -38.40) >pau.414 5804012 120 + 6537648 UUAAGUUGGGAGGAAGGGCAGUAAGUUAAUACCUUGCUGUUUUGACGUUACCAACAGAAUAAGCACCGGCUAACUUCGUGCCAGCAGCCGCGGUAAUACGAAGGGUGCAAGCGUUAAUCG ....(((((..(..(((((((((((.......)))))))))))..)....))))).......(((((......(((((((((.((....)))))...)))))))))))............ ( -38.40) >pss.3053 5952634 120 - 6093698 UUAAGUUGGGAGGAAGGGCAGUUACCUAAUACGUGAUUGUUUUGACGUUACCGACAGAAUAAGCACCGGCUAACUCUGUGCCAGCAGCCGCGGUAAUACAGAGGGUGCAAGCGUUAAUCG ....(((((..(..(((((((((((.......)))))))))))..)....))))).......(((((......(((((((((.((....)))))...)))))))))))............ ( -36.80) >pf5.74 6951530 120 + 7074893 UUAAGUUGGGAGGAAGGGCAGUUACCUAAUACGUGAUUGUUUUGACGUUACCGACAGAAUAAGCACCGGCUAACUCUGUGCCAGCAGCCGCGGUAAUACAGAGGGUGCAAGCGUUAAUCG ....(((((..(..(((((((((((.......)))))))))))..)....))))).......(((((......(((((((((.((....)))))...)))))))))))............ ( -36.80) >pfo.66 6316388 120 + 6438405 UUAAGUUGGGAGGAAGGGUUGUAGAUUAAUACUCUGCAAUUUUGACGUUACCGACAGAAUAAGCACCGGCUAACUCUGUGCCAGCAGCCGCGGUAAUACAGAGGGUGCAAGCGUUAAUCG ....(((((..(..(((((((((((.......)))))))))))..)....))))).......(((((......(((((((((.((....)))))...)))))))))))............ ( -37.50) >psp.2973 5783811 120 - 5928787 UUAAGUUGGGAGGAAGGGCAGUUACCUAAUACGUGAUUGUUUUGACGUUACCGACAGAAUAAGCACCGGCUAACUCUGUGCCAGCAGCCGCGGUAAUACAGAGGGUGCAAGCGUUAAUCG ....(((((..(..(((((((((((.......)))))))))))..)....))))).......(((((......(((((((((.((....)))))...)))))))))))............ ( -36.80) >consensus UUAAGUUGGGAGGAAGGGCAGUAACCUAAUACGUGACUGUUUUGACGUUACCGACAGAAUAAGCACCGGCUAACUCUGUGCCAGCAGCCGCGGUAAUACAGAGGGUGCAAGCGUUAAUCG ....(((((..(..(((((((((((.......)))))))))))..)....))))).......(((((......(((((((((.((....)))))...)))))))))))............ (-41.00 = -37.45 + -3.55)
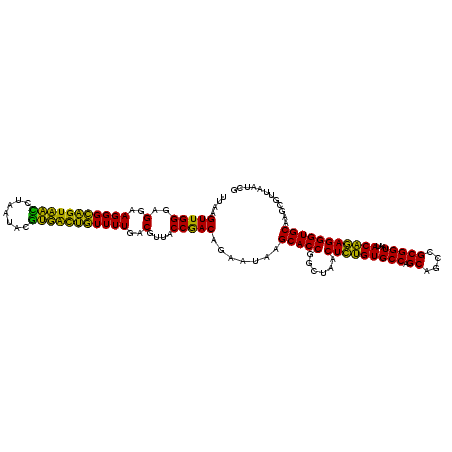
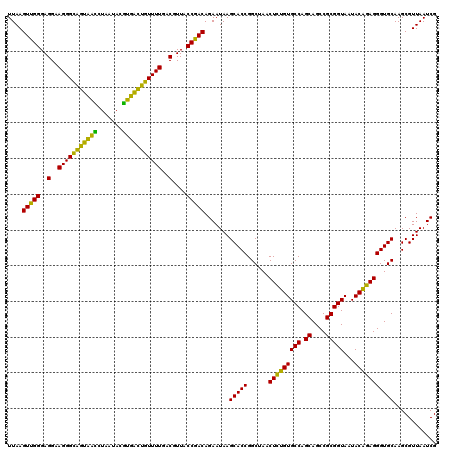
| Location | 4,793,221 – 4,793,341 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.28 |
| Mean single sequence MFE | -38.75 |
| Consensus MFE | -41.41 |
| Energy contribution | -38.75 |
| Covariance contribution | -2.66 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.05 |
| Structure conservation index | 1.07 |
| SVM decision value | 5.30 |
| SVM RNA-class probability | 0.999982 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
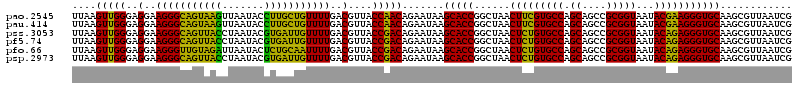
>pao.2545 4793221 120 - 6264404 UGCCGCGUGUGUGAAGAAGGUCUUCGGAUUGUAAAGCACUUUAAGUUGGGAGGAAGGGCAGUAAGUUAAUACCUUGCUGUUUUGACGUUACCAACAGAAUAAGCACCGGCUAACUUCGUG .((((.(((((((.....((((....))))......))).....(((((..(..(((((((((((.......)))))))))))..)....))))).......)))))))).......... ( -40.30) >pau.414 5804052 120 + 6537648 UGCCGCGUGUGUGAAGAAGGUCUUCGGAUUGUAAAGCACUUUAAGUUGGGAGGAAGGGCAGUAAGUUAAUACCUUGCUGUUUUGACGUUACCAACAGAAUAAGCACCGGCUAACUUCGUG .((((.(((((((.....((((....))))......))).....(((((..(..(((((((((((.......)))))))))))..)....))))).......)))))))).......... ( -40.30) >pss.3053 5952674 120 - 6093698 UGCCGCGUGUGUGAAGAAGGUCUUCGGAUUGUAAAGCACUUUAAGUUGGGAGGAAGGGCAGUUACCUAAUACGUGAUUGUUUUGACGUUACCGACAGAAUAAGCACCGGCUAACUCUGUG .((((.(((((((.....((((....))))......))).....(((((..(..(((((((((((.......)))))))))))..)....))))).......)))))))).......... ( -37.80) >pf5.74 6951570 120 + 7074893 UGCCGCGUGUGUGAAGAAGGUCUUCGGAUUGUAAAGCACUUUAAGUUGGGAGGAAGGGCAGUUACCUAAUACGUGAUUGUUUUGACGUUACCGACAGAAUAAGCACCGGCUAACUCUGUG .((((.(((((((.....((((....))))......))).....(((((..(..(((((((((((.......)))))))))))..)....))))).......)))))))).......... ( -37.80) >pfo.66 6316428 120 + 6438405 UGCCGCGUGUGUGAAGAAGGUCUUCGGAUUGUAAAGCACUUUAAGUUGGGAGGAAGGGUUGUAGAUUAAUACUCUGCAAUUUUGACGUUACCGACAGAAUAAGCACCGGCUAACUCUGUG .((((.(((((((.....((((....))))......))).....(((((..(..(((((((((((.......)))))))))))..)....))))).......)))))))).......... ( -38.50) >psp.2973 5783851 120 - 5928787 UGCCGCGUGUGUGAAGAAGGUCUUCGGAUUGUAAAGCACUUUAAGUUGGGAGGAAGGGCAGUUACCUAAUACGUGAUUGUUUUGACGUUACCGACAGAAUAAGCACCGGCUAACUCUGUG .((((.(((((((.....((((....))))......))).....(((((..(..(((((((((((.......)))))))))))..)....))))).......)))))))).......... ( -37.80) >consensus UGCCGCGUGUGUGAAGAAGGUCUUCGGAUUGUAAAGCACUUUAAGUUGGGAGGAAGGGCAGUAACCUAAUACGUGACUGUUUUGACGUUACCGACAGAAUAAGCACCGGCUAACUCUGUG .((((.(((((((.....((((....))))......))).....(((((..(..(((((((((((.......)))))))))))..)....))))).......)))))))).......... (-41.41 = -38.75 + -2.66)

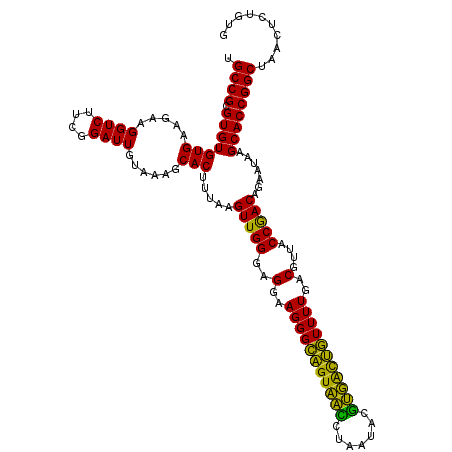
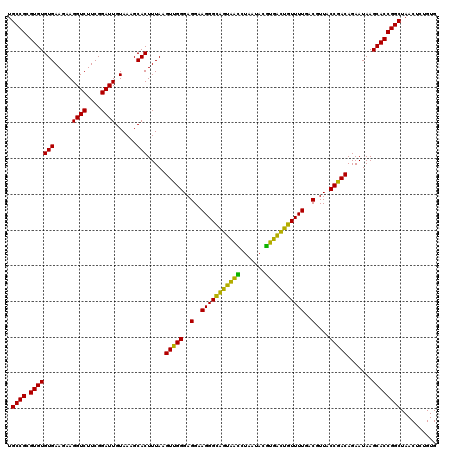
| Location | 4,793,261 – 4,793,381 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.61 |
| Mean single sequence MFE | -33.77 |
| Consensus MFE | -35.39 |
| Energy contribution | -32.95 |
| Covariance contribution | -2.44 |
| Combinations/Pair | 1.24 |
| Mean z-score | -0.78 |
| Structure conservation index | 1.05 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932219 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4793261 120 - 6264404 UGGGGAAUAUUGGACAAUGGGCGAAAGCCUGAUCCAGCCAUGCCGCGUGUGUGAAGAAGGUCUUCGGAUUGUAAAGCACUUUAAGUUGGGAGGAAGGGCAGUAAGUUAAUACCUUGCUGU .........(((((...(((((....))))).)))))((.(.((((..((((......((((....)))).....)))).....).))).)))....((((((((.......)))))))) ( -34.30) >pss.3053 5952714 120 - 6093698 UGGGGAAUAUUGGACAAUGGGCGAAAGCCUGAUCCAGCCAUGCCGCGUGUGUGAAGAAGGUCUUCGGAUUGUAAAGCACUUUAAGUUGGGAGGAAGGGCAGUUACCUAAUACGUGAUUGU ((((......((((...(((((....))))).))))(((.(.((((..((((......((((....)))).....)))).....).))).).....))).....))))............ ( -32.80) >ppk.338 5483586 120 + 6181863 UGGGGAAUAUUGGACAAUGGGCGAAAGCCUGAUCCAGCCAUGCCGCGUGUGUGAAGAAGGUCUUCGGAUUGUAAAGCACUUUAAGUUGGGAGGAAGGGCAGUAAGUUAAUACCUUGCUGU .........(((((...(((((....))))).)))))((.(.((((..((((......((((....)))).....)))).....).))).)))....((((((((.......)))))))) ( -34.30) >pf5.74 6951610 120 + 7074893 UGGGGAAUAUUGGACAAUGGGCGAAAGCCUGAUCCAGCCAUGCCGCGUGUGUGAAGAAGGUCUUCGGAUUGUAAAGCACUUUAAGUUGGGAGGAAGGGCAGUUACCUAAUACGUGAUUGU ((((......((((...(((((....))))).))))(((.(.((((..((((......((((....)))).....)))).....).))).).....))).....))))............ ( -32.80) >pfo.66 6316468 120 + 6438405 UGGGGAAUAUUGGACAAUGGGCGAAAGCCUGAUCCAGCCAUGCCGCGUGUGUGAAGAAGGUCUUCGGAUUGUAAAGCACUUUAAGUUGGGAGGAAGGGUUGUAGAUUAAUACUCUGCAAU ((.(((.((((((.(...((((....))))(((((..((.(.((((..((((......((((....)))).....)))).....).))).)))..)))))...).)))))).))).)).. ( -35.60) >psp.2973 5783891 120 - 5928787 UGGGGAAUAUUGGACAAUGGGCGAAAGCCUGAUCCAGCCAUGCCGCGUGUGUGAAGAAGGUCUUCGGAUUGUAAAGCACUUUAAGUUGGGAGGAAGGGCAGUUACCUAAUACGUGAUUGU ((((......((((...(((((....))))).))))(((.(.((((..((((......((((....)))).....)))).....).))).).....))).....))))............ ( -32.80) >consensus UGGGGAAUAUUGGACAAUGGGCGAAAGCCUGAUCCAGCCAUGCCGCGUGUGUGAAGAAGGUCUUCGGAUUGUAAAGCACUUUAAGUUGGGAGGAAGGGCAGUAACCUAAUACGUGACUGU .........(((((...(((((....))))).)))))((.(.((((..((((......((((....)))).....)))).....).))).)))....((((((((.......)))))))) (-35.39 = -32.95 + -2.44)

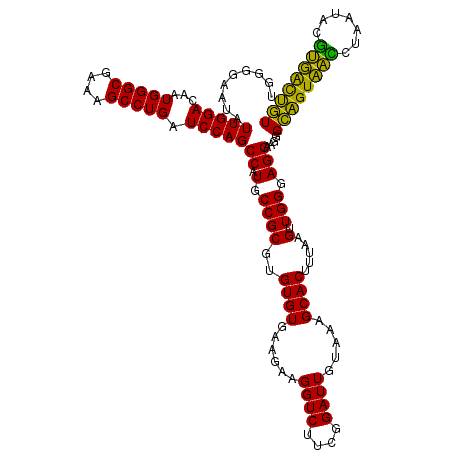
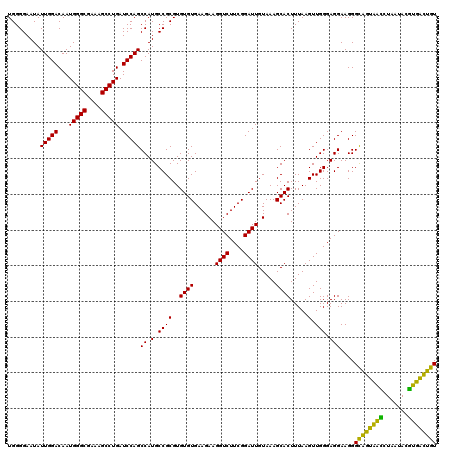
| Location | 4,793,301 – 4,793,421 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -32.80 |
| Consensus MFE | -32.80 |
| Energy contribution | -32.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.81 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.798539 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4793301 120 + 6264404 AGUGCUUUACAAUCCGAAGACCUUCUUCACACACGCGGCAUGGCUGGAUCAGGCUUUCGCCCAUUGUCCAAUAUUCCCCACUGCUGCCUCCCGUAGGAGUCUGGACCGUGUCUCAGUUCC ...............((((.....))))......(.(((((((.(((((..(((....)))....)))))....(((..(((.((((.....)))).)))..)))))))))).)...... ( -32.80) >pel.2704 5003259 120 + 5888780 AGUGCUUUACAAUCCGAAGACCUUCUUCACACACGCGGCAUGGCUGGAUCAGGCUUUCGCCCAUUGUCCAAUAUUCCCCACUGCUGCCUCCCGUAGGAGUCUGGACCGUGUCUCAGUUCC ...............((((.....))))......(.(((((((.(((((..(((....)))....)))))....(((..(((.((((.....)))).)))..)))))))))).)...... ( -32.80) >ppk.338 5483626 120 - 6181863 AGUGCUUUACAAUCCGAAGACCUUCUUCACACACGCGGCAUGGCUGGAUCAGGCUUUCGCCCAUUGUCCAAUAUUCCCCACUGCUGCCUCCCGUAGGAGUCUGGACCGUGUCUCAGUUCC ...............((((.....))))......(.(((((((.(((((..(((....)))....)))))....(((..(((.((((.....)))).)))..)))))))))).)...... ( -32.80) >pf5.74 6951650 120 - 7074893 AGUGCUUUACAAUCCGAAGACCUUCUUCACACACGCGGCAUGGCUGGAUCAGGCUUUCGCCCAUUGUCCAAUAUUCCCCACUGCUGCCUCCCGUAGGAGUCUGGACCGUGUCUCAGUUCC ...............((((.....))))......(.(((((((.(((((..(((....)))....)))))....(((..(((.((((.....)))).)))..)))))))))).)...... ( -32.80) >pfo.66 6316508 120 - 6438405 AGUGCUUUACAAUCCGAAGACCUUCUUCACACACGCGGCAUGGCUGGAUCAGGCUUUCGCCCAUUGUCCAAUAUUCCCCACUGCUGCCUCCCGUAGGAGUCUGGACCGUGUCUCAGUUCC ...............((((.....))))......(.(((((((.(((((..(((....)))....)))))....(((..(((.((((.....)))).)))..)))))))))).)...... ( -32.80) >psp.2973 5783931 120 + 5928787 AGUGCUUUACAAUCCGAAGACCUUCUUCACACACGCGGCAUGGCUGGAUCAGGCUUUCGCCCAUUGUCCAAUAUUCCCCACUGCUGCCUCCCGUAGGAGUCUGGACCGUGUCUCAGUUCC ...............((((.....))))......(.(((((((.(((((..(((....)))....)))))....(((..(((.((((.....)))).)))..)))))))))).)...... ( -32.80) >consensus AGUGCUUUACAAUCCGAAGACCUUCUUCACACACGCGGCAUGGCUGGAUCAGGCUUUCGCCCAUUGUCCAAUAUUCCCCACUGCUGCCUCCCGUAGGAGUCUGGACCGUGUCUCAGUUCC ...............((((.....))))......(.(((((((.(((((..(((....)))....)))))....(((..(((.((((.....)))).)))..)))))))))).)...... (-32.80 = -32.80 + 0.00)

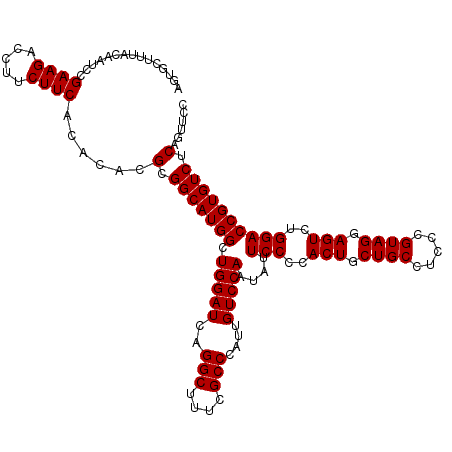
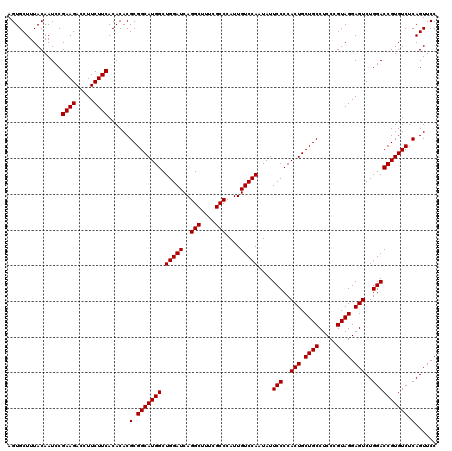
| Location | 4,793,301 – 4,793,421 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -42.70 |
| Consensus MFE | -42.70 |
| Energy contribution | -42.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.67 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.99 |
| SVM RNA-class probability | 0.998050 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4793301 120 - 6264404 GGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGGACAAUGGGCGAAAGCCUGAUCCAGCCAUGCCGCGUGUGUGAAGAAGGUCUUCGGAUUGUAAAGCACU (((.(((.....))))))...((((....))))((.(((.(((......(((((...(((((....))))).))))))))))).))..((((......((((....)))).....)))). ( -42.70) >pel.2704 5003259 120 - 5888780 GGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGGACAAUGGGCGAAAGCCUGAUCCAGCCAUGCCGCGUGUGUGAAGAAGGUCUUCGGAUUGUAAAGCACU (((.(((.....))))))...((((....))))((.(((.(((......(((((...(((((....))))).))))))))))).))..((((......((((....)))).....)))). ( -42.70) >ppk.338 5483626 120 + 6181863 GGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGGACAAUGGGCGAAAGCCUGAUCCAGCCAUGCCGCGUGUGUGAAGAAGGUCUUCGGAUUGUAAAGCACU (((.(((.....))))))...((((....))))((.(((.(((......(((((...(((((....))))).))))))))))).))..((((......((((....)))).....)))). ( -42.70) >pf5.74 6951650 120 + 7074893 GGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGGACAAUGGGCGAAAGCCUGAUCCAGCCAUGCCGCGUGUGUGAAGAAGGUCUUCGGAUUGUAAAGCACU (((.(((.....))))))...((((....))))((.(((.(((......(((((...(((((....))))).))))))))))).))..((((......((((....)))).....)))). ( -42.70) >pfo.66 6316508 120 + 6438405 GGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGGACAAUGGGCGAAAGCCUGAUCCAGCCAUGCCGCGUGUGUGAAGAAGGUCUUCGGAUUGUAAAGCACU (((.(((.....))))))...((((....))))((.(((.(((......(((((...(((((....))))).))))))))))).))..((((......((((....)))).....)))). ( -42.70) >psp.2973 5783931 120 - 5928787 GGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGGACAAUGGGCGAAAGCCUGAUCCAGCCAUGCCGCGUGUGUGAAGAAGGUCUUCGGAUUGUAAAGCACU (((.(((.....))))))...((((....))))((.(((.(((......(((((...(((((....))))).))))))))))).))..((((......((((....)))).....)))). ( -42.70) >consensus GGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGGACAAUGGGCGAAAGCCUGAUCCAGCCAUGCCGCGUGUGUGAAGAAGGUCUUCGGAUUGUAAAGCACU (((.(((.....))))))...((((....))))((.(((.(((......(((((...(((((....))))).))))))))))).))..((((......((((....)))).....)))). (-42.70 = -42.70 + -0.00)
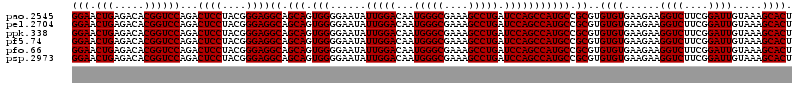
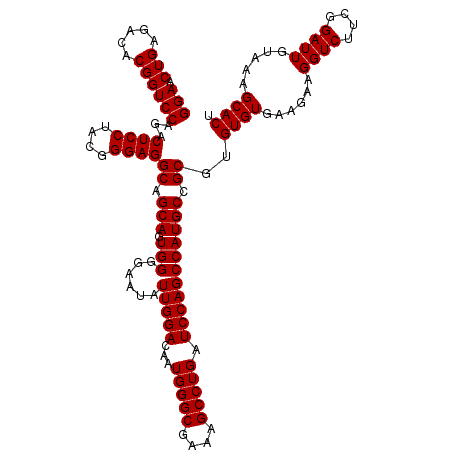
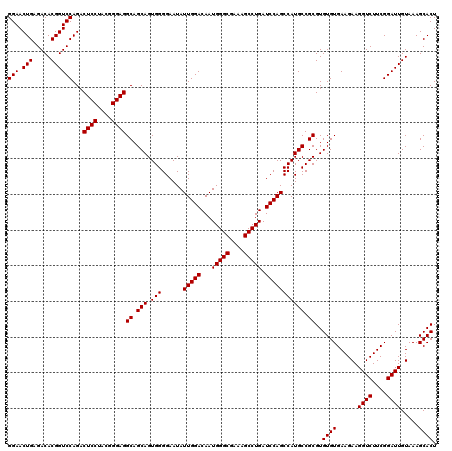
| Location | 4,793,341 – 4,793,461 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -43.50 |
| Consensus MFE | -43.50 |
| Energy contribution | -43.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.09 |
| Structure conservation index | 1.00 |
| SVM decision value | 3.90 |
| SVM RNA-class probability | 0.999695 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4793341 120 - 6264404 GACGAUCCGUAACUGGUCUGAGAGGAUGAUCAGUCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGGACAAUGGGCGAAAGCCUGAUCCAGCCA .....(((...(((((((.........)))))))(((.(((((.(((.....)))))))).((((....))))......))).)))...(((((...(((((....))))).)))))... ( -43.50) >pel.2704 5003299 120 - 5888780 GACGAUCCGUAACUGGUCUGAGAGGAUGAUCAGUCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGGACAAUGGGCGAAAGCCUGAUCCAGCCA .....(((...(((((((.........)))))))(((.(((((.(((.....)))))))).((((....))))......))).)))...(((((...(((((....))))).)))))... ( -43.50) >ppk.338 5483666 120 + 6181863 GACGAUCCGUAACUGGUCUGAGAGGAUGAUCAGUCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGGACAAUGGGCGAAAGCCUGAUCCAGCCA .....(((...(((((((.........)))))))(((.(((((.(((.....)))))))).((((....))))......))).)))...(((((...(((((....))))).)))))... ( -43.50) >pf5.74 6951690 120 + 7074893 GACGAUCCGUAACUGGUCUGAGAGGAUGAUCAGUCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGGACAAUGGGCGAAAGCCUGAUCCAGCCA .....(((...(((((((.........)))))))(((.(((((.(((.....)))))))).((((....))))......))).)))...(((((...(((((....))))).)))))... ( -43.50) >pfo.66 6316548 120 + 6438405 GACGAUCCGUAACUGGUCUGAGAGGAUGAUCAGUCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGGACAAUGGGCGAAAGCCUGAUCCAGCCA .....(((...(((((((.........)))))))(((.(((((.(((.....)))))))).((((....))))......))).)))...(((((...(((((....))))).)))))... ( -43.50) >psp.2973 5783971 120 - 5928787 GACGAUCCGUAACUGGUCUGAGAGGAUGAUCAGUCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGGACAAUGGGCGAAAGCCUGAUCCAGCCA .....(((...(((((((.........)))))))(((.(((((.(((.....)))))))).((((....))))......))).)))...(((((...(((((....))))).)))))... ( -43.50) >consensus GACGAUCCGUAACUGGUCUGAGAGGAUGAUCAGUCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGGACAAUGGGCGAAAGCCUGAUCCAGCCA .....(((...(((((((.........)))))))(((.(((((.(((.....)))))))).((((....))))......))).)))...(((((...(((((....))))).)))))... (-43.50 = -43.50 + 0.00)

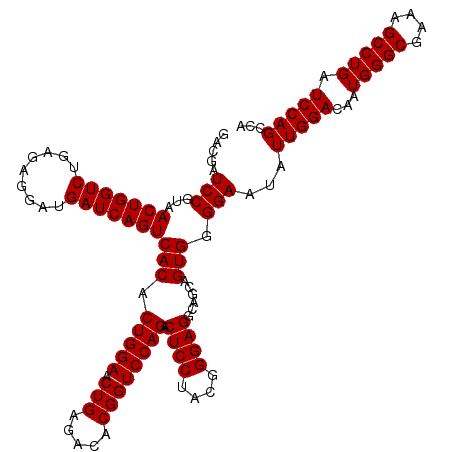
| Location | 4,793,381 – 4,793,501 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.22 |
| Mean single sequence MFE | -46.73 |
| Consensus MFE | -47.40 |
| Energy contribution | -46.73 |
| Covariance contribution | -0.67 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.41 |
| Structure conservation index | 1.01 |
| SVM decision value | 5.09 |
| SVM RNA-class probability | 0.999973 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4793381 120 - 6264404 GUCGGAUUAGCUAGUUGGUGGGGUAAAGGCCUACCAAGGCGACGAUCCGUAACUGGUCUGAGAGGAUGAUCAGUCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAG ...(((((.(((..((((((((.......)))))))))))...)))))((.(((((((.........))))))).)).(((((.(((.....)))))))).((((....))))....... ( -46.90) >pau.414 5804212 120 + 6537648 GUCGGAUUAGCUAGUUGGUGGGGUAAAGGCCUACCAAGGCGACGAUCCGUAACUGGUCUGAGAGGAUGAUCAGUCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAG ...(((((.(((..((((((((.......)))))))))))...)))))((.(((((((.........))))))).)).(((((.(((.....)))))))).((((....))))....... ( -46.90) >pel.2704 5003339 120 - 5888780 GUCGGAUUAGCUAGUUGGUGGGGUAAUGGCUCACCAAGGCGACGAUCCGUAACUGGUCUGAGAGGAUGAUCAGUCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAG ...(((((.(((..((((((((.......)))))))))))...)))))((.(((((((.........))))))).)).(((((.(((.....)))))))).((((....))))....... ( -46.20) >ppk.338 5483706 120 + 6181863 GUCGGAUUAGCUAGUUGGUGGGGUAAUGGCUCACCAAGGCGACGAUCCGUAACUGGUCUGAGAGGAUGAUCAGUCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAG ...(((((.(((..((((((((.......)))))))))))...)))))((.(((((((.........))))))).)).(((((.(((.....)))))))).((((....))))....... ( -46.20) >pfo.66 6316588 120 + 6438405 GUCGGAUUAGCUAGUUGGUGAGGUAAUGGCUCACCAAGGCGACGAUCCGUAACUGGUCUGAGAGGAUGAUCAGUCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAG ...(((((.(((..((((((((.......)))))))))))...)))))((.(((((((.........))))))).)).(((((.(((.....)))))))).((((....))))....... ( -47.10) >psp.2973 5784011 120 - 5928787 GUCGGAUUAGCUAGUUGGUGAGGUAAUGGCUCACCAAGGCGACGAUCCGUAACUGGUCUGAGAGGAUGAUCAGUCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAG ...(((((.(((..((((((((.......)))))))))))...)))))((.(((((((.........))))))).)).(((((.(((.....)))))))).((((....))))....... ( -47.10) >consensus GUCGGAUUAGCUAGUUGGUGGGGUAAUGGCUCACCAAGGCGACGAUCCGUAACUGGUCUGAGAGGAUGAUCAGUCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAG ...(((((.(((..((((((((.......)))))))))))...)))))((.(((((((.........))))))).)).(((((.(((.....)))))))).((((....))))....... (-47.40 = -46.73 + -0.67)

| Location | 4,793,421 – 4,793,541 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.94 |
| Mean single sequence MFE | -34.69 |
| Consensus MFE | -32.17 |
| Energy contribution | -32.03 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.846553 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4793421 120 + 6264404 AGUGUGACUGAUCAUCCUCUCAGACCAGUUACGGAUCGUCGCCUUGGUAGGCCUUUACCCCACCAACUAGCUAAUCCGACCUAGGCUCAUCUGAUAGCGUGAGGUCCGAAGAUCCCCCAC ...(((((((.((.........)).)))))))(((((...((.(((((.((.......)).)))))...))...((.((((((.(((........))).).))))).)).)))))..... ( -34.60) >pau.414 5804252 120 - 6537648 AGUGUGACUGAUCAUCCUCUCAGACCAGUUACGGAUCGUCGCCUUGGUAGGCCUUUACCCCACCAACUAGCUAAUCCGACCUAGGCUCAUCUGAUAGCGUGAGGUCCGAAGAUCCCCCAC ...(((((((.((.........)).)))))))(((((...((.(((((.((.......)).)))))...))...((.((((((.(((........))).).))))).)).)))))..... ( -34.60) >pel.2704 5003379 120 + 5888780 AGUGUGACUGAUCAUCCUCUCAGACCAGUUACGGAUCGUCGCCUUGGUGAGCCAUUACCCCACCAACUAGCUAAUCCGACCUAGGCUCAUCUGAUAGCGCAAGGCCCGAAGGUCCCCUGC ...(((((((.((.........)).)))))))(((((.(((((((((((((((..............................))))))).........))))))..)).)))))..... ( -32.21) >ppk.338 5483746 120 - 6181863 AGUGUGACUGAUCAUCCUCUCAGACCAGUUACGGAUCGUCGCCUUGGUGAGCCAUUACCCCACCAACUAGCUAAUCCGACCUAGGCUCAUCUGAUAGCGCAAGGCCCGAAGGUCCCCUGC ...(((((((.((.........)).)))))))(((((.(((((((((((((((..............................))))))).........))))))..)).)))))..... ( -32.21) >pf5.74 6951770 120 - 7074893 AGUGUGACUGAUCAUCCUCUCAGACCAGUUACGGAUCGUCGCCUUGGUGAGCCAUUACCUCACCAACUAGCUAAUCCGACCUAGGCUCAUCUAAUAGCGUGAGGUCCGAAGAUCCCCCAC ...(((((((.((.........)).)))))))(((((...((.((((((((.......))))))))...))...((.((((((.(((........))).).))))).)).)))))..... ( -39.00) >psp.2973 5784051 120 + 5928787 AGUGUGACUGAUCAUCCUCUCAGACCAGUUACGGAUCGUCGCCUUGGUGAGCCAUUACCUCACCAACUAGCUAAUCCGACCUAGGCUCAUCUGAUAGCGCAAGGCCCGAAGGUCCCCUGC ...(((((((.((.........)).)))))))(((((.(((((((((((((.......)))))).....((((..((......)).((....))))))..)))))..)).)))))..... ( -35.50) >consensus AGUGUGACUGAUCAUCCUCUCAGACCAGUUACGGAUCGUCGCCUUGGUGAGCCAUUACCCCACCAACUAGCUAAUCCGACCUAGGCUCAUCUGAUAGCGCAAGGCCCGAAGAUCCCCCAC ...(((((((.((.........)).)))))))(((((.(((((((((((((.......)))))).....((((.((.((..........)).))))))..)))))..)).)))))..... (-32.17 = -32.03 + -0.14)
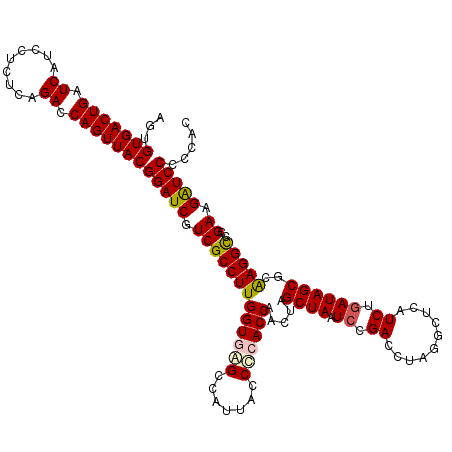
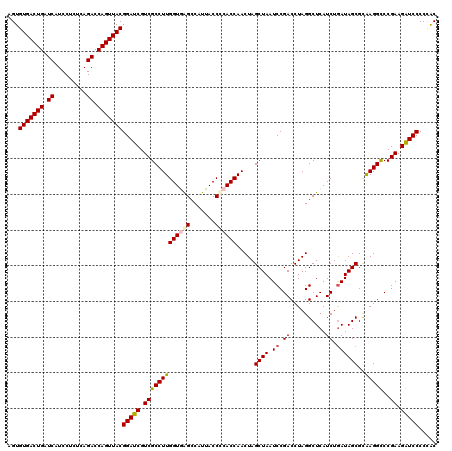
| Location | 4,793,421 – 4,793,541 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.94 |
| Mean single sequence MFE | -44.08 |
| Consensus MFE | -45.92 |
| Energy contribution | -43.62 |
| Covariance contribution | -2.30 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.44 |
| Structure conservation index | 1.04 |
| SVM decision value | 3.41 |
| SVM RNA-class probability | 0.999161 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4793421 120 - 6264404 GUGGGGGAUCUUCGGACCUCACGCUAUCAGAUGAGCCUAGGUCGGAUUAGCUAGUUGGUGGGGUAAAGGCCUACCAAGGCGACGAUCCGUAACUGGUCUGAGAGGAUGAUCAGUCACACU ((((..((((((((((((....(((........)))..((..((((((.(((..((((((((.......)))))))))))...))))))...)))))))))).....))))..))))... ( -43.50) >pau.414 5804252 120 + 6537648 GUGGGGGAUCUUCGGACCUCACGCUAUCAGAUGAGCCUAGGUCGGAUUAGCUAGUUGGUGGGGUAAAGGCCUACCAAGGCGACGAUCCGUAACUGGUCUGAGAGGAUGAUCAGUCACACU ((((..((((((((((((....(((........)))..((..((((((.(((..((((((((.......)))))))))))...))))))...)))))))))).....))))..))))... ( -43.50) >pel.2704 5003379 120 - 5888780 GCAGGGGACCUUCGGGCCUUGCGCUAUCAGAUGAGCCUAGGUCGGAUUAGCUAGUUGGUGGGGUAAUGGCUCACCAAGGCGACGAUCCGUAACUGGUCUGAGAGGAUGAUCAGUCACACU ((((((..((....)))))))).((.(((((.....((((..((((((.(((..((((((((.......)))))))))))...))))))...))))))))).))(((.....)))..... ( -44.30) >ppk.338 5483746 120 + 6181863 GCAGGGGACCUUCGGGCCUUGCGCUAUCAGAUGAGCCUAGGUCGGAUUAGCUAGUUGGUGGGGUAAUGGCUCACCAAGGCGACGAUCCGUAACUGGUCUGAGAGGAUGAUCAGUCACACU ((((((..((....)))))))).((.(((((.....((((..((((((.(((..((((((((.......)))))))))))...))))))...))))))))).))(((.....)))..... ( -44.30) >pf5.74 6951770 120 + 7074893 GUGGGGGAUCUUCGGACCUCACGCUAUUAGAUGAGCCUAGGUCGGAUUAGCUAGUUGGUGAGGUAAUGGCUCACCAAGGCGACGAUCCGUAACUGGUCUGAGAGGAUGAUCAGUCACACU ((((..((((((((((((....(((........)))..((..((((((.(((..((((((((.......)))))))))))...))))))...)))))))))).....))))..))))... ( -43.70) >psp.2973 5784051 120 - 5928787 GCAGGGGACCUUCGGGCCUUGCGCUAUCAGAUGAGCCUAGGUCGGAUUAGCUAGUUGGUGAGGUAAUGGCUCACCAAGGCGACGAUCCGUAACUGGUCUGAGAGGAUGAUCAGUCACACU ((((((..((....)))))))).((.(((((.....((((..((((((.(((..((((((((.......)))))))))))...))))))...))))))))).))(((.....)))..... ( -45.20) >consensus GCAGGGGACCUUCGGACCUCACGCUAUCAGAUGAGCCUAGGUCGGAUUAGCUAGUUGGUGGGGUAAUGGCUCACCAAGGCGACGAUCCGUAACUGGUCUGAGAGGAUGAUCAGUCACACU ((((((..((....)))))))).((.(((((.....((((..((((((.(((..((((((((.......)))))))))))...))))))...))))))))).))(((.....)))..... (-45.92 = -43.62 + -2.30)

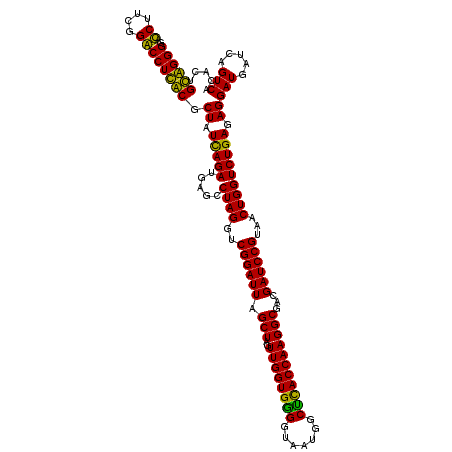
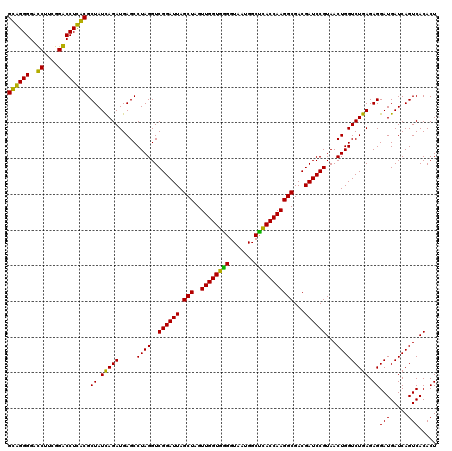
| Location | 4,793,461 – 4,793,581 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.06 |
| Mean single sequence MFE | -42.23 |
| Consensus MFE | -43.42 |
| Energy contribution | -40.75 |
| Covariance contribution | -2.67 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.15 |
| Structure conservation index | 1.03 |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.981765 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4793461 120 - 6264404 GGAAACGGGCGCUAAUACCGCAUACGUCCUGAGGGAGAAAGUGGGGGAUCUUCGGACCUCACGCUAUCAGAUGAGCCUAGGUCGGAUUAGCUAGUUGGUGGGGUAAAGGCCUACCAAGGC (....)..((((((((.(((....((((.(((...((...((((((..((....)))))))).)).))))))).((....)))))))))))...((((((((.......)))))))).)) ( -41.90) >pau.414 5804292 120 + 6537648 GGAAACGGGCGCUAAUACCGCAUACGUCCUGAGGGAGAAAGUGGGGGAUCUUCGGACCUCACGCUAUCAGAUGAGCCUAGGUCGGAUUAGCUAGUUGGUGGGGUAAAGGCCUACCAAGGC (....)..((((((((.(((....((((.(((...((...((((((..((....)))))))).)).))))))).((....)))))))))))...((((((((.......)))))))).)) ( -41.90) >pel.2704 5003419 120 - 5888780 CGAAAGGAACGCUAAUACCGCAUACGUCCUACGGGAGAAAGCAGGGGACCUUCGGGCCUUGCGCUAUCAGAUGAGCCUAGGUCGGAUUAGCUAGUUGGUGGGGUAAUGGCUCACCAAGGC (....)....((((((.((((.....(((....)))((.(((.((((.((....))))))..))).))............).)))))))))...((((((((.......))))))))... ( -42.00) >ppk.338 5483786 120 + 6181863 CGAAAGGAACGCUAAUACCGCAUACGUCCUACGGGAGAAAGCAGGGGACCUUCGGGCCUUGCGCUAUCAGAUGAGCCUAGGUCGGAUUAGCUAGUUGGUGGGGUAAUGGCUCACCAAGGC (....)....((((((.((((.....(((....)))((.(((.((((.((....))))))..))).))............).)))))))))...((((((((.......))))))))... ( -42.00) >pf5.74 6951810 120 + 7074893 GGAAACGGGCGCUAAUACCGCAUACGUCCUACGGGAGAAAGUGGGGGAUCUUCGGACCUCACGCUAUUAGAUGAGCCUAGGUCGGAUUAGCUAGUUGGUGAGGUAAUGGCUCACCAAGGC (....)((((.((((((..((.....(((....)))....((((((..((....))))))))))))))))....))))...........(((..((((((((.......))))))))))) ( -42.70) >pfo.66 6316668 120 + 6438405 CGAAAGGAACGCUAAUACCGCAUACGUCCUACGGGAGAAAGCAGGGGACCUUCGGGCCUUGCGCUAUCAGAUGAGCCUAGGUCGGAUUAGCUAGUUGGUGAGGUAAUGGCUCACCAAGGC (....)....((((((.((((.....(((....)))((.(((.((((.((....))))))..))).))............).)))))))))...((((((((.......))))))))... ( -42.90) >consensus CGAAACGAACGCUAAUACCGCAUACGUCCUACGGGAGAAAGCAGGGGACCUUCGGACCUCACGCUAUCAGAUGAGCCUAGGUCGGAUUAGCUAGUUGGUGGGGUAAUGGCUCACCAAGGC (....)....((((((.((((.....(((....)))....((((((..((....))))))))(((........)))....).)))))))))...((((((((.......))))))))... (-43.42 = -40.75 + -2.67)
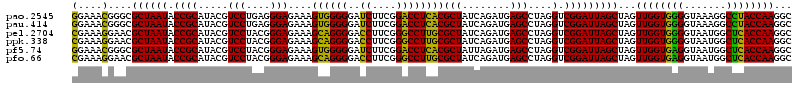
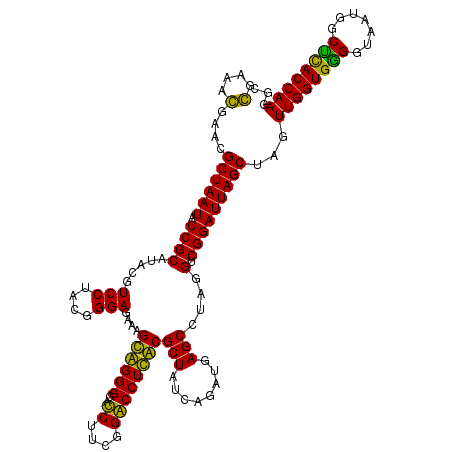
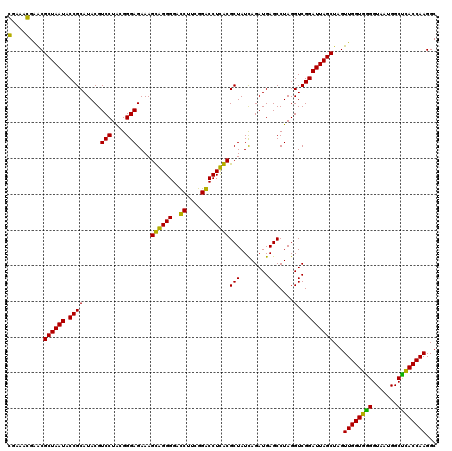
| Location | 4,793,501 – 4,793,621 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.06 |
| Mean single sequence MFE | -47.23 |
| Consensus MFE | -49.94 |
| Energy contribution | -46.67 |
| Covariance contribution | -3.28 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.71 |
| Structure conservation index | 1.06 |
| SVM decision value | 6.07 |
| SVM RNA-class probability | 0.999996 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4793501 120 - 6264404 UAAUGCCUAGGAAUCUGCCUGGUAGUGGGGGAUAACGUCCGGAAACGGGCGCUAAUACCGCAUACGUCCUGAGGGAGAAAGUGGGGGAUCUUCGGACCUCACGCUAUCAGAUGAGCCUAG ......(((((..((...(((((((((((((....((((((....))))))......((.(((...(((....)))....))).))..((....)))))).)))))))))..)).))))) ( -47.60) >pau.414 5804332 120 + 6537648 UAAUGCCUAGGAAUCUGCCUGGUAGUGGGGGAUAACGUCCGGAAACGGGCGCUAAUACCGCAUACGUCCUGAGGGAGAAAGUGGGGGAUCUUCGGACCUCACGCUAUCAGAUGAGCCUAG ......(((((..((...(((((((((((((....((((((....))))))......((.(((...(((....)))....))).))..((....)))))).)))))))))..)).))))) ( -47.60) >pel.2704 5003459 120 - 5888780 UAAUGCCUAGGAAUCUGCCUGGUAGUGGGGGACAACGUUUCGAAAGGAACGCUAAUACCGCAUACGUCCUACGGGAGAAAGCAGGGGACCUUCGGGCCUUGCGCUAUCAGAUGAGCCUAG ......(((((.(((((..((((.(((.((.....((((((....))))))......)).)))...(((....)))....((((((..((....)))))))))))).)))))...))))) ( -46.60) >ppk.338 5483826 120 + 6181863 UAAUGCCUAGGAAUCUGCCUGGUAGUGGGGGACAACGUUUCGAAAGGAACGCUAAUACCGCAUACGUCCUACGGGAGAAAGCAGGGGACCUUCGGGCCUUGCGCUAUCAGAUGAGCCUAG ......(((((.(((((..((((.(((.((.....((((((....))))))......)).)))...(((....)))....((((((..((....)))))))))))).)))))...))))) ( -46.60) >pf5.74 6951850 120 + 7074893 UAAUGCCUAGGAAUCUGCCUAGUAGUGGGGGAUAACGUCCGGAAACGGGCGCUAAUACCGCAUACGUCCUACGGGAGAAAGUGGGGGAUCUUCGGACCUCACGCUAUUAGAUGAGCCUAG ......(((((..((...(((((((((((((....((((((....))))))......((.(((...(((....)))....))).))..((....)))))).)))))))))..)).))))) ( -48.40) >pfo.66 6316708 120 + 6438405 UAAUGCCUAGGAAUCUGCCUGGUAGUGGGGGACAACGUUUCGAAAGGAACGCUAAUACCGCAUACGUCCUACGGGAGAAAGCAGGGGACCUUCGGGCCUUGCGCUAUCAGAUGAGCCUAG ......(((((.(((((..((((.(((.((.....((((((....))))))......)).)))...(((....)))....((((((..((....)))))))))))).)))))...))))) ( -46.60) >consensus UAAUGCCUAGGAAUCUGCCUGGUAGUGGGGGACAACGUCCCGAAACGAACGCUAAUACCGCAUACGUCCUACGGGAGAAAGCAGGGGACCUUCGGACCUCACGCUAUCAGAUGAGCCUAG ......(((((.(((((..((((.(((.((.....((((((....))))))......)).)))...(((....)))....((((((..((....)))))))))))).)))))...))))) (-49.94 = -46.67 + -3.28)
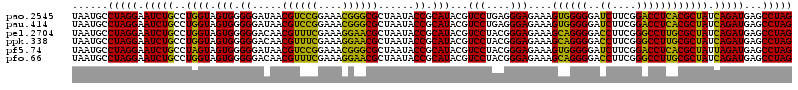

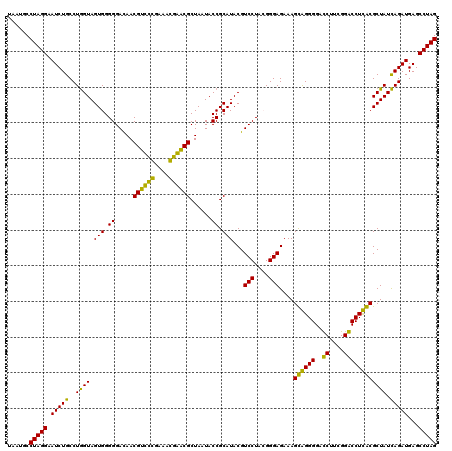
| Location | 4,793,541 – 4,793,660 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.37 |
| Mean single sequence MFE | -43.63 |
| Consensus MFE | -41.07 |
| Energy contribution | -39.52 |
| Covariance contribution | -1.55 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.940130 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4793541 119 - 6264404 AAGGGAGCUUGCUCCUGGAUUC-AGCGGCGGACGGGUGAGUAAUGCCUAGGAAUCUGCCUGGUAGUGGGGGAUAACGUCCGGAAACGGGCGCUAAUACCGCAUACGUCCUGAGGGAGAAA ..(((((....)))))...(((-((.((((.............((((.(((......)))))))(((.((..((.((((((....)))))).))...)).))).)))))))))....... ( -44.30) >pau.414 5804372 119 + 6537648 AAGGGAGCUUGCUCCUGGAUUC-AGCGGCGGACGGGUGAGUAAUGCCUAGGAAUCUGCCUGGUAGUGGGGGAUAACGUCCGGAAACGGGCGCUAAUACCGCAUACGUCCUGAGGGAGAAA ..(((((....)))))...(((-((.((((.............((((.(((......)))))))(((.((..((.((((((....)))))).))...)).))).)))))))))....... ( -44.30) >pss.3053 5952994 120 - 6093698 ACGGGUACUUGUACCUGGUGGCGAGCGGCGGACGGGUGAGUAAUGCCUAGGAAUCUGCCUGGUAGUGGGGGAUAACGCUCGGAAACGGACGCUAAUACCGCAUACGUCCUACGGGAGAAA .((((((....))))))((((..(((((((...((((.......))))....((((.((.......)).))))..)))(((....))).))))....)))).....(((....))).... ( -42.30) >pf5.74 6951890 120 + 7074893 ACGGGUACUUGUACCUGGUGGCGAGCGGCGGACGGGUGAGUAAUGCCUAGGAAUCUGCCUAGUAGUGGGGGAUAACGUCCGGAAACGGGCGCUAAUACCGCAUACGUCCUACGGGAGAAA .((((((....))))))((((..((((.(((((((((.......)))).(..((((.((.......)).))))..)))))(....).).))))....)))).....(((....))).... ( -46.40) >pfo.66 6316748 118 + 6438405 A-GGGAGCUUGCUCCUGGAUUC-AGCGGCGGACGGGUGAGUAAUGCCUAGGAAUCUGCCUGGUAGUGGGGGACAACGUUUCGAAAGGAACGCUAAUACCGCAUACGUCCUACGGGAGAAA .-(((((....)))))...(((-.((((((((.((((.......)))).....)))))).((((...(....)..((((((....))))))....)))))).....(((....)))))). ( -42.20) >psp.2973 5784171 120 - 5928787 ACGGGUACUUGUACCUGGUGGCGAGCGGCGGACGGGUGAGUAAUGCCUAGGAAUCUGCCUGGUAGUGGGGGAUAACGCUCGGAAACGGACGCUAAUACCGCAUACGUCCUACGGGAGAAA .((((((....))))))((((..(((((((...((((.......))))....((((.((.......)).))))..)))(((....))).))))....)))).....(((....))).... ( -42.30) >consensus ACGGGAACUUGCACCUGGAGGC_AGCGGCGGACGGGUGAGUAAUGCCUAGGAAUCUGCCUGGUAGUGGGGGAUAACGUCCGGAAACGGACGCUAAUACCGCAUACGUCCUACGGGAGAAA ..(((((....)))))..........((((((.((((.......)))).....))))))..(((((((.......((((((....))))))......)))).))).(((....))).... (-41.07 = -39.52 + -1.55)

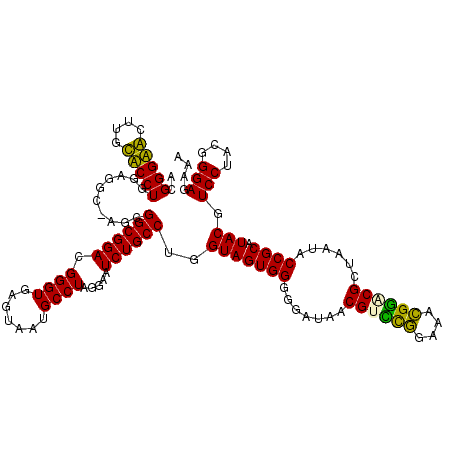
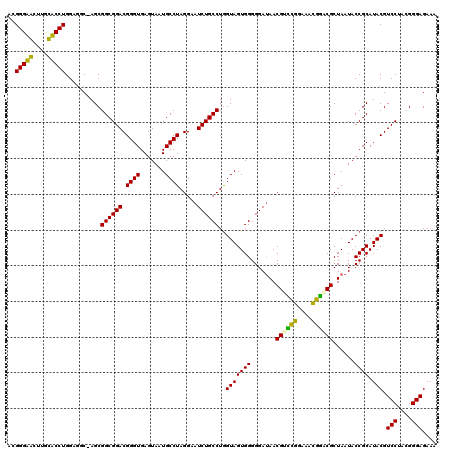
Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:10:19 2007