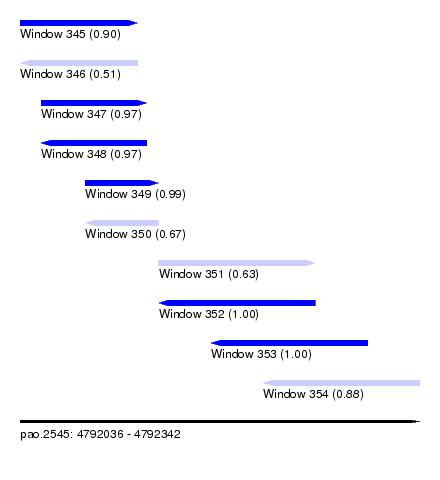
| Sequence ID | pao.2545 |
|---|---|
| Location | 4,792,036 – 4,792,342 |
| Length | 306 |
| Max. P | 0.999874 |

| Location | 4,792,036 – 4,792,126 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 85.26 |
| Mean single sequence MFE | -31.46 |
| Consensus MFE | -26.82 |
| Energy contribution | -26.60 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.903048 |
| Prediction | RNA |
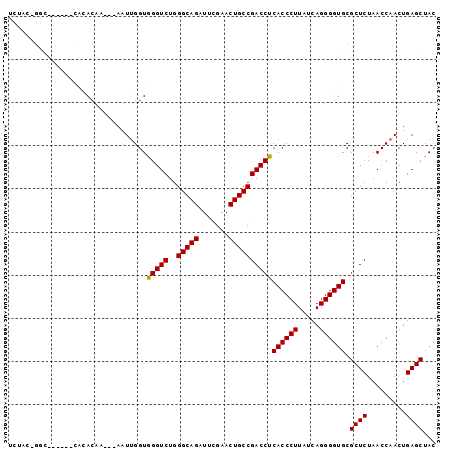
Download alignment: ClustalW | MAF
>pao.2545 4792036 90 + 6264404 CACGCAG--------CACACCAAC-AAUUGGUGGGUCUGGGCAGAUUCGAACUGCCGACCUCACCCUUAUCAGGGGUGCGCUCUAACCAACUGAGCUAC ......(--------(((.((...-....((((((((..(((((.......))))))))).))))........))))))((((.........))))... ( -33.36) >pel.2704 5001970 91 + 5888780 UCUA--GGC------CGCACCAAGUAAUUGGUAGGUCUGGGCAGAUUUGAACUGCCGACCUCACCCUUAUCAGGGGUGCGCUCUAACCAACUGAGCUAC ..((--(((------(..(((((....))))).))))))(((((.......))))).....((((((.....)))))).((((.........))))... ( -36.20) >ppk.338 5482338 91 - 6181863 UCUAC-GGC------UGAACCAUGUAAU-GGUAGGUCUGGGCAGAUUUGAACUGCCGACCUCACCCUUAUCAGGGGUGCGCUCUAACCAACUGAGCUAC .....-...------...(((((...))-)))(((((..(((((.......))))))))))((((((.....)))))).((((.........))))... ( -29.60) >pf5.74 6950337 78 - 7074893 ------------------ACAA---AAUUGGUGGGUCUGGGCAGAUUCGAACUGCCGACCUCACCCUUAUCAGGGGUGCGCUCUAACCAACUGAGCUAC ------------------....---.......(((((..(((((.......))))))))))((((((.....)))))).((((.........))))... ( -26.60) >pfo.66 6315219 96 - 6438405 UCUACAGGCGUUUCCCACACAA---AAUUGGUGGGUCUGGGCAGAUUCGAACUGCCGACCUCACCCUUAUCAGGGGUGCGCUCUAACCAACUGAGCUAC ....((((.....(((((.((.---...)))))))))))(((((.......))))).....((((((.....)))))).((((.........))))... ( -31.50) >psp.2973 5782600 96 + 5928787 UCUACAGGCGUUUCCCACACAA---AAUUGGUGGGUCUGGGCAGAUUCGAACUGCCGACCUCACCCUUAUCAGGGGUGCGCUCUAACCAACUGAGCUAC ....((((.....(((((.((.---...)))))))))))(((((.......))))).....((((((.....)))))).((((.........))))... ( -31.50) >consensus UCUAC_GGC______CACACAA___AAUUGGUGGGUCUGGGCAGAUUCGAACUGCCGACCUCACCCUUAUCAGGGGUGCGCUCUAACCAACUGAGCUAC ................................(((((..(((((.......))))))))))((((((.....)))))).((((.........))))... (-26.82 = -26.60 + -0.22)

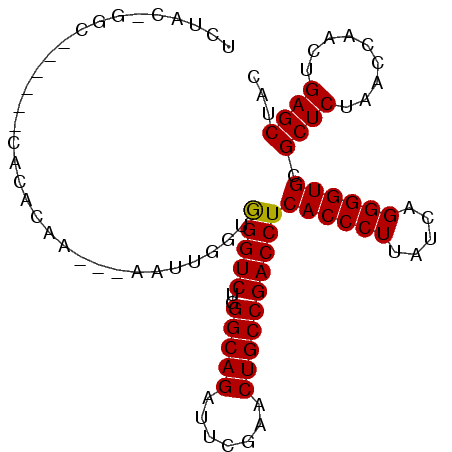
| Location | 4,792,036 – 4,792,126 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 85.26 |
| Mean single sequence MFE | -31.87 |
| Consensus MFE | -26.20 |
| Energy contribution | -26.31 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.82 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.511960 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4792036 90 - 6264404 GUAGCUCAGUUGGUUAGAGCGCACCCCUGAUAAGGGUGAGGUCGGCAGUUCGAAUCUGCCCAGACCCACCAAUU-GUUGGUGUG--------CUGCGUG (((((((.........))))((((.((.(((((.((((.(((((((((.......)))))..))))))))..))-))))).)))--------))))... ( -36.30) >pel.2704 5001970 91 - 5888780 GUAGCUCAGUUGGUUAGAGCGCACCCCUGAUAAGGGUGAGGUCGGCAGUUCAAAUCUGCCCAGACCUACCAAUUACUUGGUGCG------GCC--UAGA ...((((.........))))(((((..((((...(((.((((((((((.......)))))..)))))))).))))...))))).------...--.... ( -32.70) >ppk.338 5482338 91 + 6181863 GUAGCUCAGUUGGUUAGAGCGCACCCCUGAUAAGGGUGAGGUCGGCAGUUCAAAUCUGCCCAGACCUACC-AUUACAUGGUUCA------GCC-GUAGA ...........((((.((((.(((((.......)))))((((((((((.......)))))..)))))...-........)))))------)))-..... ( -30.60) >pf5.74 6950337 78 + 7074893 GUAGCUCAGUUGGUUAGAGCGCACCCCUGAUAAGGGUGAGGUCGGCAGUUCGAAUCUGCCCAGACCCACCAAUU---UUGU------------------ ...((..(((((((.......(((((.......))))).(((((((((.......)))))..)))).)))))))---..))------------------ ( -26.80) >pfo.66 6315219 96 + 6438405 GUAGCUCAGUUGGUUAGAGCGCACCCCUGAUAAGGGUGAGGUCGGCAGUUCGAAUCUGCCCAGACCCACCAAUU---UUGUGUGGGAAACGCCUGUAGA ...((((.........)))).(((((.......)))))((((.(((((.......)))))....(((((((...---.)).)))))....))))..... ( -32.40) >psp.2973 5782600 96 - 5928787 GUAGCUCAGUUGGUUAGAGCGCACCCCUGAUAAGGGUGAGGUCGGCAGUUCGAAUCUGCCCAGACCCACCAAUU---UUGUGUGGGAAACGCCUGUAGA ...((((.........)))).(((((.......)))))((((.(((((.......)))))....(((((((...---.)).)))))....))))..... ( -32.40) >consensus GUAGCUCAGUUGGUUAGAGCGCACCCCUGAUAAGGGUGAGGUCGGCAGUUCGAAUCUGCCCAGACCCACCAAUU___UGGUGUG______GCC_GUAGA ...((((.........))))((((..........((((.(((((((((.......)))))..)))))))).........))))................ (-26.20 = -26.31 + 0.11)

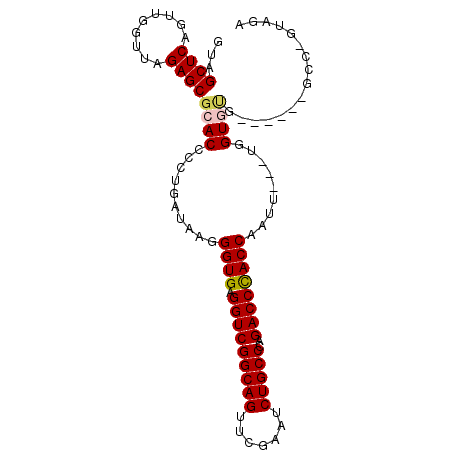
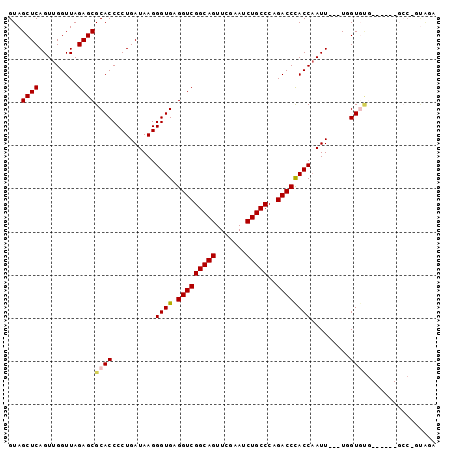
| Location | 4,792,052 – 4,792,133 |
|---|---|
| Length | 81 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.51 |
| Mean single sequence MFE | -39.08 |
| Consensus MFE | -35.90 |
| Energy contribution | -35.48 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.60 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970362 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
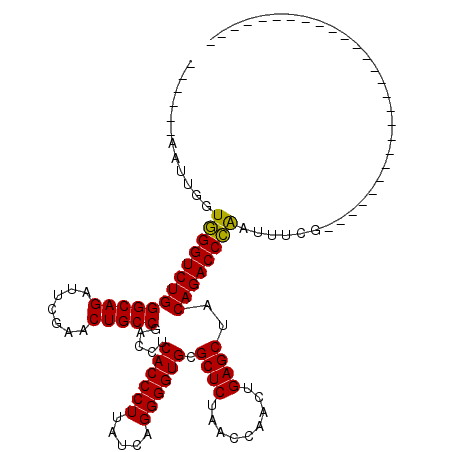
>pao.2545 4792052 81 + 6264404 ------AAUUGGUGGGUCUGGGCAGAUUCGAACUGCCGACCUCACCCUUAUCAGGGGUGCGCUCUAACCAACUGAGCUACAGACCCA--------------------------------- ------......(((((((((((((.......))))).....((((((.....)))))).((((.........))))..))))))))--------------------------------- ( -35.30) >pss.3053 5951479 96 + 6093698 CCAGUAAACUGGUGGGUCUGGGCAGAUUCGAACUGCCGACCUCACCCUUAUCAGGGGUGCGCUCUAACCAACUGAGCUACAGACCCGAACUCGGGC------------------------ ((((....))))(((((((((((((.......))))).....((((((.....)))))).((((.........))))..)))))))).........------------------------ ( -39.80) >ppk.338 5482356 86 - 6181863 ------AAU-GGUAGGUCUGGGCAGAUUUGAACUGCCGACCUCACCCUUAUCAGGGGUGCGCUCUAACCAACUGAGCUACAGACCUAUAACAG--------------------------- ------...-.((((((((((((((.......))))).....((((((.....)))))).((((.........))))..))))))))).....--------------------------- ( -35.60) >pf5.74 6950341 114 - 7074893 ------AAUUGGUGGGUCUGGGCAGAUUCGAACUGCCGACCUCACCCUUAUCAGGGGUGCGCUCUAACCAACUGAGCUACAGACCCAAUUUCGAGCUUGUAACUGUUAGCUUGGAGCUAU ------...((((((((((((((((.......))))).....((((((.....)))))).((((.........))))..)))))))...((((((((..........)))))))))))). ( -44.90) >pfo.66 6315241 87 - 6438405 ------AAUUGGUGGGUCUGGGCAGAUUCGAACUGCCGACCUCACCCUUAUCAGGGGUGCGCUCUAACCAACUGAGCUACAGACCCAAUUUCG--------------------------- ------......(((((((((((((.......))))).....((((((.....)))))).((((.........))))..))))))))......--------------------------- ( -36.10) >psp.2973 5782622 114 + 5928787 ------AAUUGGUGGGUCUGGGCAGAUUCGAACUGCCGACCUCACCCUUAUCAGGGGUGCGCUCUAACCAACUGAGCUACAGACCCAAUUUCGAGCUUGUAGCCGCUAGCGUUGAGCUAU ------......(((((((((((((.......))))).....((((((.....)))))).((((.........))))..))))))))......(((((...((.....))...))))).. ( -42.80) >consensus ______AAUUGGUGGGUCUGGGCAGAUUCGAACUGCCGACCUCACCCUUAUCAGGGGUGCGCUCUAACCAACUGAGCUACAGACCCAAUUUCG___________________________ ............(((((((((((((.......))))).....((((((.....)))))).((((.........))))..))))))))................................. (-35.90 = -35.48 + -0.42)
| Location | 4,792,052 – 4,792,133 |
|---|---|
| Length | 81 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.51 |
| Mean single sequence MFE | -38.37 |
| Consensus MFE | -33.31 |
| Energy contribution | -33.03 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.92 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974361 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
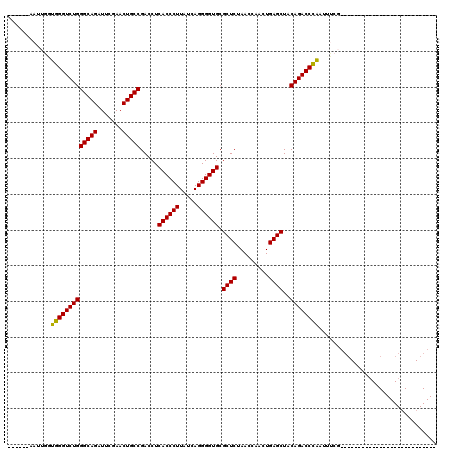
>pao.2545 4792052 81 - 6264404 ---------------------------------UGGGUCUGUAGCUCAGUUGGUUAGAGCGCACCCCUGAUAAGGGUGAGGUCGGCAGUUCGAAUCUGCCCAGACCCACCAAUU------ ---------------------------------(((((((((((((.....)))))((.(.(((((.......))))).).))(((((.......)))))))))))))......------ ( -33.80) >pss.3053 5951479 96 - 6093698 ------------------------GCCCGAGUUCGGGUCUGUAGCUCAGUUGGUUAGAGCGCACCCCUGAUAAGGGUGAGGUCGGCAGUUCGAAUCUGCCCAGACCCACCAGUUUACUGG ------------------------((((....(((((..(((.((((.........)))))))..)))))...))))(.(((((((((.......)))))..))))).((((....)))) ( -38.60) >ppk.338 5482356 86 + 6181863 ---------------------------CUGUUAUAGGUCUGUAGCUCAGUUGGUUAGAGCGCACCCCUGAUAAGGGUGAGGUCGGCAGUUCAAAUCUGCCCAGACCUACC-AUU------ ---------------------------......(((((((((((((.....)))))((.(.(((((.......))))).).))(((((.......)))))))))))))..-...------ ( -32.10) >pf5.74 6950341 114 + 7074893 AUAGCUCCAAGCUAACAGUUACAAGCUCGAAAUUGGGUCUGUAGCUCAGUUGGUUAGAGCGCACCCCUGAUAAGGGUGAGGUCGGCAGUUCGAAUCUGCCCAGACCCACCAAUU------ ...((((..((((((((((((((..((((....))))..)))))))..))))))).))))..............((((.(((((((((.......)))))..))))))))....------ ( -45.00) >pfo.66 6315241 87 + 6438405 ---------------------------CGAAAUUGGGUCUGUAGCUCAGUUGGUUAGAGCGCACCCCUGAUAAGGGUGAGGUCGGCAGUUCGAAUCUGCCCAGACCCACCAAUU------ ---------------------------......(((((((((((((.....)))))((.(.(((((.......))))).).))(((((.......)))))))))))))......------ ( -34.00) >psp.2973 5782622 114 - 5928787 AUAGCUCAACGCUAGCGGCUACAAGCUCGAAAUUGGGUCUGUAGCUCAGUUGGUUAGAGCGCACCCCUGAUAAGGGUGAGGUCGGCAGUUCGAAUCUGCCCAGACCCACCAAUU------ ...((((...(((((((((((((..((((....))))..)))))))..))))))..))))..............((((.(((((((((.......)))))..))))))))....------ ( -46.70) >consensus ___________________________CGAAAUUGGGUCUGUAGCUCAGUUGGUUAGAGCGCACCCCUGAUAAGGGUGAGGUCGGCAGUUCGAAUCUGCCCAGACCCACCAAUU______ ..................................((((((((((((.....)))))((.(.(((((.......))))).).))(((((.......))))))))))))............. (-33.31 = -33.03 + -0.28)

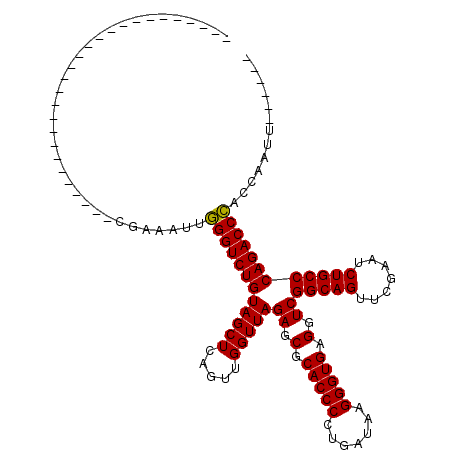
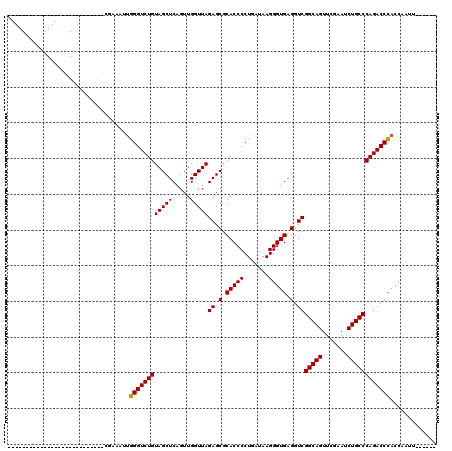
| Location | 4,792,086 – 4,792,142 |
|---|---|
| Length | 56 |
| Sequences | 6 |
| Columns | 81 |
| Reading direction | forward |
| Mean pairwise identity | 75.73 |
| Mean single sequence MFE | -19.85 |
| Consensus MFE | -13.55 |
| Energy contribution | -13.55 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.68 |
| SVM decision value | 2.32 |
| SVM RNA-class probability | 0.992285 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
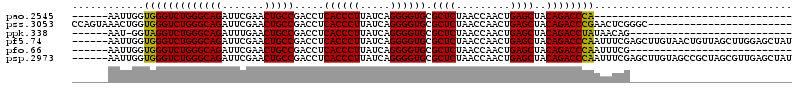
>pao.2545 4792086 56 + 6264404 CUCACCCUUAUCAGGGGUGCGCUCUAACCAACUGAGCUACAGACCCA-----------------------AUCGUCUAA-- ..((((((.....)))))).((((.........))))...((((...-----------------------...))))..-- ( -16.20) >pau.414 5802917 56 - 6537648 CUCACCCUUAUCAGGGGUGCGCUCUAACCAACUGAGCUACAGACCCA-----------------------AUCGUCUAA-- ..((((((.....)))))).((((.........))))...((((...-----------------------...))))..-- ( -16.20) >pss.3053 5951519 76 + 6093698 CUCACCCUUAUCAGGGGUGCGCUCUAACCAACUGAGCUACAGACCCGAACUCGGGC-GCUGCUUCUA---AUCGUCUUC-U ..((((((.....)))))).((((.........))))..(((.((((....)))).-.)))......---.........-. ( -22.50) >pel.2704 5002021 78 + 5888780 CUCACCCUUAUCAGGGGUGCGCUCUAACCAACUGAGCUACAGACCUAUAACAG---GGUCGCGUUACAGCAUCGUCUUUAC ..((((((.....)))))).((((.........))))....(((((......)---))))((......))........... ( -22.70) >ppk.338 5482389 77 - 6181863 CUCACCCUUAUCAGGGGUGCGCUCUAACCAACUGAGCUACAGACCUAUAACAG---GGUAGCGUUACAGCAUCGUCUUU-U ..((((((.....)))))).(((.(((((....).(((((...(((.....))---)))))))))).))).........-. ( -20.60) >pfo.66 6315275 73 - 6438405 CUCACCCUUAUCAGGGGUGCGCUCUAACCAACUGAGCUACAGACCCAAUUUCG---GGCUGCUUCUU----UCGUCUUC-U ..((((((.....)))))).((((.........))))..(((.(((......)---)))))......----........-. ( -20.90) >consensus CUCACCCUUAUCAGGGGUGCGCUCUAACCAACUGAGCUACAGACCCA_A___G____G__GC_U______AUCGUCUUA_U ..((((((.....)))))).((((.........))))...((((.............................)))).... (-13.55 = -13.55 + -0.00)

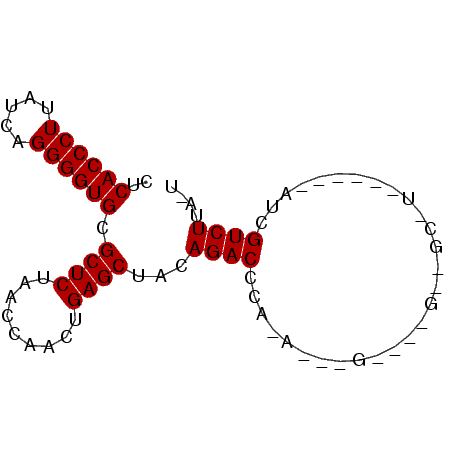
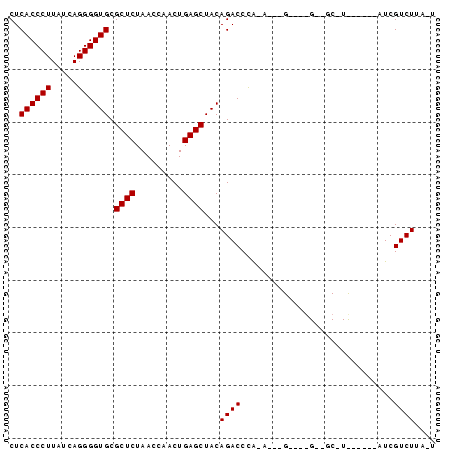
| Location | 4,792,086 – 4,792,142 |
|---|---|
| Length | 56 |
| Sequences | 6 |
| Columns | 81 |
| Reading direction | reverse |
| Mean pairwise identity | 75.73 |
| Mean single sequence MFE | -21.25 |
| Consensus MFE | -13.32 |
| Energy contribution | -13.32 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.671430 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4792086 56 - 6264404 --UUAGACGAU-----------------------UGGGUCUGUAGCUCAGUUGGUUAGAGCGCACCCCUGAUAAGGGUGAG --.....((((-----------------------(((((.....))))))))).........(((((.......))))).. ( -16.60) >pau.414 5802917 56 + 6537648 --UUAGACGAU-----------------------UGGGUCUGUAGCUCAGUUGGUUAGAGCGCACCCCUGAUAAGGGUGAG --.....((((-----------------------(((((.....))))))))).........(((((.......))))).. ( -16.60) >pss.3053 5951519 76 - 6093698 A-GAAGACGAU---UAGAAGCAGC-GCCCGAGUUCGGGUCUGUAGCUCAGUUGGUUAGAGCGCACCCCUGAUAAGGGUGAG .-.....((((---(((..((((.-(((((....)))))))))..)).))))).........(((((.......))))).. ( -25.10) >pel.2704 5002021 78 - 5888780 GUAAAGACGAUGCUGUAACGCGACC---CUGUUAUAGGUCUGUAGCUCAGUUGGUUAGAGCGCACCCCUGAUAAGGGUGAG ...........(((.((((.((((.---..((((((....))))))...)))))))).))).(((((.......))))).. ( -24.80) >ppk.338 5482389 77 + 6181863 A-AAAGACGAUGCUGUAACGCUACC---CUGUUAUAGGUCUGUAGCUCAGUUGGUUAGAGCGCACCCCUGAUAAGGGUGAG .-.........(((.((((((((((---(((...))))...))))).(....))))).))).(((((.......))))).. ( -21.50) >pfo.66 6315275 73 + 6438405 A-GAAGACGA----AAGAAGCAGCC---CGAAAUUGGGUCUGUAGCUCAGUUGGUUAGAGCGCACCCCUGAUAAGGGUGAG .-.....(..----..)..((((((---((....)))).)))).((((.........)))).(((((.......))))).. ( -22.90) >consensus A_AAAGACGAU______A_GC__C____C___U_UGGGUCUGUAGCUCAGUUGGUUAGAGCGCACCCCUGAUAAGGGUGAG ....((((.............................))))...((((.........)))).(((((.......))))).. (-13.32 = -13.32 + 0.00)

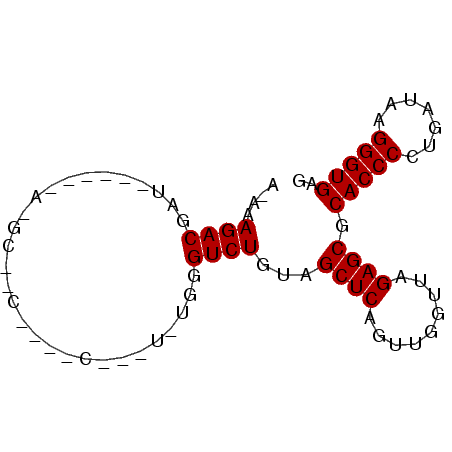
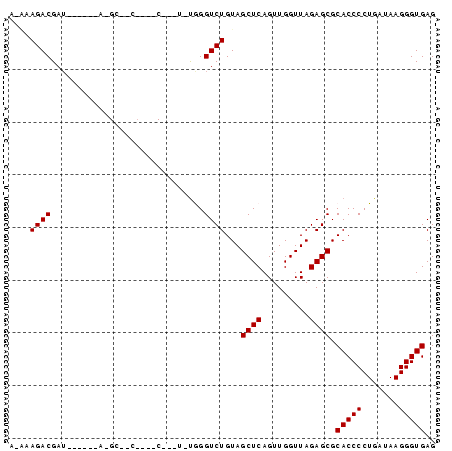
| Location | 4,792,142 – 4,792,262 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.50 |
| Mean single sequence MFE | -37.60 |
| Consensus MFE | -34.35 |
| Energy contribution | -34.80 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.627000 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4792142 120 + 6264404 CCAGUGAAUCAAGCAAUUCGUGUGGGAGCUUAUGAAGAAGCUGAGAUCUUCGAUUAAGGAGGUGAUCCAGCCGCAGGUUCCCCUACGGCUACCUUGUUACGACUUCACCCCAGUCAUGAA ................(((((((((((((((......)))))..((...(((..(((((.((....))(((((.(((....))).))))).)))))...)))..))..))))..)))))) ( -35.10) >pau.414 5802973 120 - 6537648 CCAGUGAAUCAAGCAAUUCGUGUGGGAGCUUAUGAAGAAGCUGAGAUCUUCGAUUAAGGAGGUGAUCCAGCCGCAGGUUCCCCUACGGCUACCUUGUUACGACUUCACCCCAGUCAUGAA ................(((((((((((((((......)))))..((...(((..(((((.((....))(((((.(((....))).))))).)))))...)))..))..))))..)))))) ( -35.10) >pss.3053 5951595 119 + 6093698 UCAAUGAAUCAAGCAAUUCGUGUGGGUGCUUAUGGUGCAGCUGA-GUCGUCGAUUAAGGAGGUGAUCCAGCCGCAGGUUCCCCUACGGCUACCUUGUUACGACUUCACCCCAGUCAUGAA ................(((((((((((((.......)))...((-(((((....(((((.((....))(((((.(((....))).))))).)))))..)))))))...))))..)))))) ( -37.30) >pel.2704 5002099 120 + 5888780 ACAAUGAAUCAAGCAAUUCGUGUGGGAGCUCAUCAGUAGGCUGAUGUCGUCGAUUAAGGAGGUGAUCCAGCCGCAGGUUCCCCUACGGCUACCUUGUUACGACUUCACCCCAGUCAUGAA ................((((((((((....((((((....))))))..((((..(((((.((....))(((((.(((....))).))))).)))))...)))).....))))..)))))) ( -40.30) >ppk.338 5482466 120 - 6181863 ACAAUGAAUCAAGCAAUUCGUGUGGGAGCUCAUCAGCAGGCUGAUGUCGUCGAUUAAGGAGGUGAUCCAGCCGCAGGUUCCCCUACGGCUACCUUGUUACGACUUCACCCCAGUCAUGAA ................((((((((((....((((((....))))))..((((..(((((.((....))(((((.(((....))).))))).)))))...)))).....))))..)))))) ( -40.50) >psp.2973 5782772 119 + 5928787 UCAAUGAAUCAAGCAAUUCGUGUGGGUGCUUAUGGUGCAGCUGA-GUCGUCGAUUAAGGAGGUGAUCCAGCCGCAGGUUCCCCUACGGCUACCUUGUUACGACUUCACCCCAGUCAUGAA ................(((((((((((((.......)))...((-(((((....(((((.((....))(((((.(((....))).))))).)))))..)))))))...))))..)))))) ( -37.30) >consensus ACAAUGAAUCAAGCAAUUCGUGUGGGAGCUUAUGAAGAAGCUGA_GUCGUCGAUUAAGGAGGUGAUCCAGCCGCAGGUUCCCCUACGGCUACCUUGUUACGACUUCACCCCAGUCAUGAA ................((((((((((.(((........)))(((.(((((....(((((.((....))(((((.(((....))).))))).)))))..))))).))).))))..)))))) (-34.35 = -34.80 + 0.45)

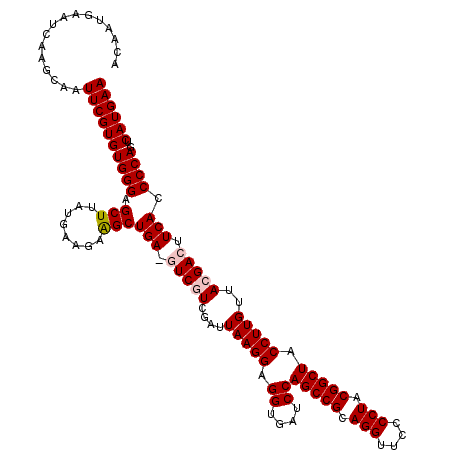
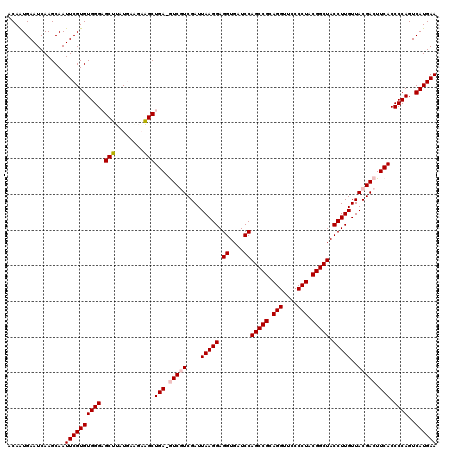
| Location | 4,792,142 – 4,792,262 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.50 |
| Mean single sequence MFE | -41.77 |
| Consensus MFE | -38.12 |
| Energy contribution | -38.57 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.67 |
| SVM RNA-class probability | 0.996236 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4792142 120 - 6264404 UUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGGGAACCUGCGGCUGGAUCACCUCCUUAAUCGAAGAUCUCAGCUUCUUCAUAAGCUCCCACACGAAUUGCUUGAUUCACUGG ..(((((..(((((((.((....))....((((((((((....))))))))))..))))))).......(((((........))))).(((((.............)))))..))).)). ( -38.02) >pau.414 5802973 120 + 6537648 UUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGGGAACCUGCGGCUGGAUCACCUCCUUAAUCGAAGAUCUCAGCUUCUUCAUAAGCUCCCACACGAAUUGCUUGAUUCACUGG ..(((((..(((((((.((....))....((((((((((....))))))))))..))))))).......(((((........))))).(((((.............)))))..))).)). ( -38.02) >pss.3053 5951595 119 - 6093698 UUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGGGAACCUGCGGCUGGAUCACCUCCUUAAUCGACGAC-UCAGCUGCACCAUAAGCACCCACACGAAUUGCUUGAUUCAUUGA ........((((.(((.(((((...((((.(((((((((....)))))))))((....)).)))).....)))))-)))(((........))).))))...(((((....)))))..... ( -42.10) >pel.2704 5002099 120 - 5888780 UUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGGGAACCUGCGGCUGGAUCACCUCCUUAAUCGACGACAUCAGCCUACUGAUGAGCUCCCACACGAAUUGCUUGAUUCAUUGU (((((.(.((((.(((.(((((...((((.(((((((((....)))))))))((....)).)))).....))))).))).)))).).))))).........(((((....)))))..... ( -45.30) >ppk.338 5482466 120 + 6181863 UUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGGGAACCUGCGGCUGGAUCACCUCCUUAAUCGACGACAUCAGCCUGCUGAUGAGCUCCCACACGAAUUGCUUGAUUCAUUGU ........((((((...((((....((((.(((((((((....)))))))))((....)).))))...))))..((((((....)))))).)).))))...(((((....)))))..... ( -45.10) >psp.2973 5782772 119 - 5928787 UUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGGGAACCUGCGGCUGGAUCACCUCCUUAAUCGACGAC-UCAGCUGCACCAUAAGCACCCACACGAAUUGCUUGAUUCAUUGA ........((((.(((.(((((...((((.(((((((((....)))))))))((....)).)))).....)))))-)))(((........))).))))...(((((....)))))..... ( -42.10) >consensus UUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGGGAACCUGCGGCUGGAUCACCUCCUUAAUCGACGAC_UCAGCUUCAUCAUAAGCUCCCACACGAAUUGCUUGAUUCAUUGA ........((((.(((.(((((...((((.(((((((((....)))))))))((....)).)))).....))))).)))(((........))).))))...(((((....)))))..... (-38.12 = -38.57 + 0.45)

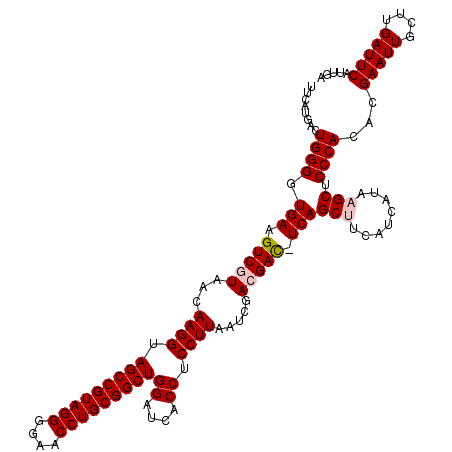
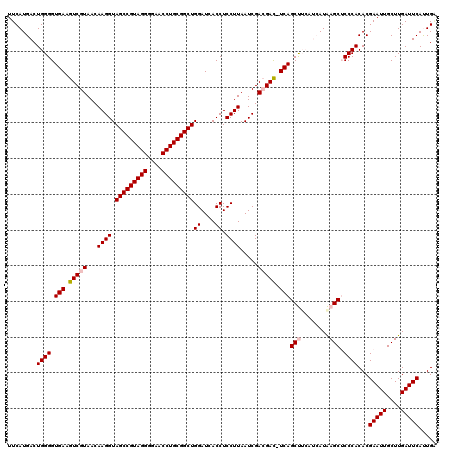
| Location | 4,792,182 – 4,792,302 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.72 |
| Mean single sequence MFE | -47.40 |
| Consensus MFE | -45.02 |
| Energy contribution | -44.80 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.95 |
| SVM decision value | 4.34 |
| SVM RNA-class probability | 0.999874 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4792182 120 - 6264404 AGCUAGUCUAACCGCAAGGGGGACGGUUACCACGGAGUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGGGAACCUGCGGCUGGAUCACCUCCUUAAUCGAAGAUCUCAG .....((((..((....)).))))(((((....((((((((((((((((.......))))))........(((((((((....))))))))))))))).)))).)))))........... ( -48.10) >pau.414 5803013 120 + 6537648 AGCUAGUCUAACCGCAAGGGGGACGGUUACCACGGAGUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGGGAACCUGCGGCUGGAUCACCUCCUUAAUCGAAGAUCUCAG .....((((..((....)).))))(((((....((((((((((((((((.......))))))........(((((((((....))))))))))))))).)))).)))))........... ( -48.10) >pss.3053 5951635 119 - 6093698 AGCUAGUCUAACCUUCGGGAGGACGGUUACCACGGUGUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGGGAACCUGCGGCUGGAUCACCUCCUUAAUCGACGAC-UCAG ....((((...((....))....((((((....((.(((((((((((((.......))))))........(((((((((....))))))))))))))))..)).))))))..)))-)... ( -46.70) >pf5.74 6950530 120 + 7074893 AGCUAGUCUAACCUUCGGGAGGACGGUUACCACGGUGUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGGGAACCUGCGGCUGGAUCACCUCCUUAAUCGACGACAUCAG ..((((((((((((((....))).))))).(((...)))......))))))((((..((((....((((.(((((((((....)))))))))((....)).))))...))))..)))).. ( -47.30) >pfo.66 6315388 120 + 6438405 AGCUAGUCUAACCUUCGGGAGGACGGUUACCACGGUGUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGGGAACCUGCGGCUGGAUCACCUCCUUAAUCGACGACAUCAG ..((((((((((((((....))).))))).(((...)))......))))))((((..((((....((((.(((((((((....)))))))))((....)).))))...))))..)))).. ( -47.30) >psp.2973 5782812 119 - 5928787 AGCUAGUCUAACCUUCGGGGGGACGGUUACCACGGUGUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGGGAACCUGCGGCUGGAUCACCUCCUUAAUCGACGAC-UCAG ....((((...((....))....((((((....((.(((((((((((((.......))))))........(((((((((....))))))))))))))))..)).))))))..)))-)... ( -46.90) >consensus AGCUAGUCUAACCUUCGGGAGGACGGUUACCACGGUGUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGGGAACCUGCGGCUGGAUCACCUCCUUAAUCGACGAC_UCAG .....(((...((....))....((((((....((.(((((((((((((.......))))))........(((((((((....))))))))))))))))))...))))))..)))..... (-45.02 = -44.80 + -0.22)

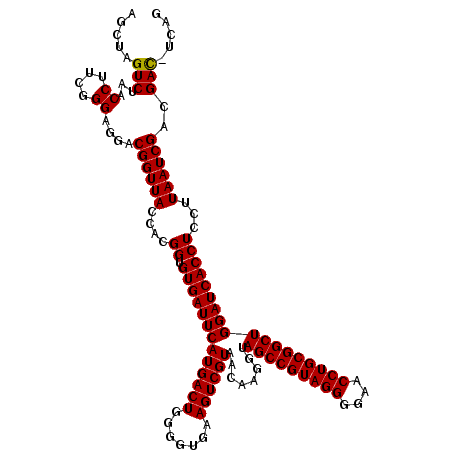
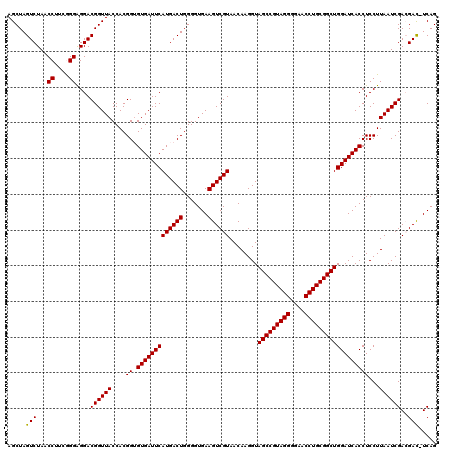
| Location | 4,792,222 – 4,792,342 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.83 |
| Mean single sequence MFE | -43.15 |
| Consensus MFE | -43.59 |
| Energy contribution | -43.15 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -0.81 |
| Structure conservation index | 1.01 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.884759 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4792222 120 - 6264404 CACCGCCCGUCACACCAUGGGAGUGGGUUGCUCCAGAAGUAGCUAGUCUAACCGCAAGGGGGACGGUUACCACGGAGUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGG .....(((.((((.((.(((..((((((..((((.(..((((((.((((..((....)).))))))))))..)))))..))))))..)))))))))...........((...))...))) ( -43.90) >pau.414 5803053 120 + 6537648 CACCGCCCGUCACACCAUGGGAGUGGGUUGCUCCAGAAGUAGCUAGUCUAACCGCAAGGGGGACGGUUACCACGGAGUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGG .....(((.((((.((.(((..((((((..((((.(..((((((.((((..((....)).))))))))))..)))))..))))))..)))))))))...........((...))...))) ( -43.90) >pss.3053 5951674 120 - 6093698 CACCGCCCGUCACACCAUGGGAGUGGGUUGCACCAGAAGUAGCUAGUCUAACCUUCGGGAGGACGGUUACCACGGUGUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGG .....(((.((((.((.(((..((((((..((((.(..((((((.((((..((....)).))))))))))..)))))..))))))..)))))))))...........((...))...))) ( -42.60) >pf5.74 6950570 120 + 7074893 CACCGCCCGUCACACCAUGGGAGUGGGUUGCACCAGAAGUAGCUAGUCUAACCUUCGGGAGGACGGUUACCACGGUGUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGG .....(((.((((.((.(((..((((((..((((.(..((((((.((((..((....)).))))))))))..)))))..))))))..)))))))))...........((...))...))) ( -42.60) >pfo.66 6315428 120 + 6438405 CACCGCCCGUCACACCAUGGGAGUGGGUUGCACCAGAAGUAGCUAGUCUAACCUUCGGGAGGACGGUUACCACGGUGUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGG .....(((.((((.((.(((..((((((..((((.(..((((((.((((..((....)).))))))))))..)))))..))))))..)))))))))...........((...))...))) ( -42.60) >psp.2973 5782851 120 - 5928787 CACCGCCCGUCACACCAUGGGAGUGGGUUGCACCAGAAGUAGCUAGUCUAACCUUCGGGGGGACGGUUACCACGGUGUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGG .....(((.((((.((.(((..((((((..((((.(..((((((.((((..((....)).))))))))))..)))))..))))))..)))))))))...........((...))...))) ( -43.30) >consensus CACCGCCCGUCACACCAUGGGAGUGGGUUGCACCAGAAGUAGCUAGUCUAACCUUCGGGAGGACGGUUACCACGGUGUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGG .....(((.((((.((.(((..((((((..((((.(..((((((.((((..((....)).))))))))))..)))))..))))))..)))))))))...........((...))...))) (-43.59 = -43.15 + -0.44)

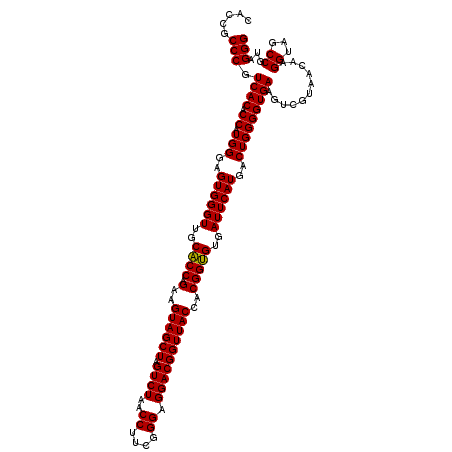
Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:09:41 2007