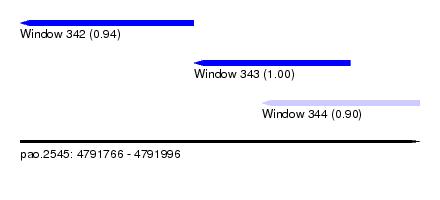
| Sequence ID | pao.2545 |
|---|---|
| Location | 4,791,766 – 4,791,996 |
| Length | 230 |
| Max. P | 0.996967 |

| Location | 4,791,766 – 4,791,866 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 77.67 |
| Mean single sequence MFE | -29.63 |
| Consensus MFE | -19.27 |
| Energy contribution | -16.78 |
| Covariance contribution | -2.49 |
| Combinations/Pair | 1.52 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.944497 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
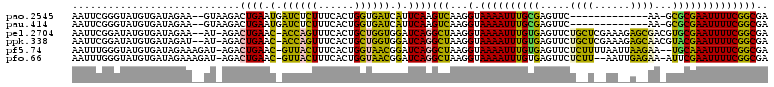
>pao.2545 4791766 100 - 6264404 AAUUCGGGUAUGUGAUAGAA--GUAAGACUGAAUGAUCUCUUUCACUGGUGAUCAUUCAAGUCAAGGUAAAAUUUGCGAGUUC-------------AA-GCGCGAAUUUUCGGCGA ...(((..............--....((((((((((((.((......)).)))))))).))))...(.((((((((((.....-------------..-.)))))))))))..))) ( -27.70) >pau.414 5802597 100 + 6537648 AAUUCGGGUAUGUGAUAGAA--GUAAGACUGAAUGAUCUCUUUCACUGGUGAUCAUUCAAGUCAAGGUAAAAUUUGCGAGUUC-------------AA-GCGCGAAUUUUCGGCGA ...(((..............--....((((((((((((.((......)).)))))))).))))...(.((((((((((.....-------------..-.)))))))))))..))) ( -27.70) >pel.2704 5001686 112 - 5888780 AAUUCGGAUAUGUGAUAGAA--AU-AGACUGAAC-ACCAGUUUCACUGCUGGUGGAUCAGGCUAAGGUAAAAUUUGUGAGUUCUGCUCGAAAGAGCGACGUGCGAAUUUUCGGCGA ...(((..............--.(-((.((((.(-((((((......)))))))..)))).)))..(.(((((((((..(((..((((....)))))))..))))))))))..))) ( -36.10) >ppk.338 5482056 112 + 6181863 AAUUCGGAUAUGUGAUAGAU--AU-AGACUGAAC-ACCAGUUUCACUGCUGGUGGAUCAGGCUAAGGUAAAAUUUGUGAGUUCUGCUCGAAAGAGCAACGUACGAAUUUUCGGCGA ...(((..............--.(-((.((((.(-((((((......)))))))..)))).)))..(.((((((((((.((..(((((....))))))).)))))))))))..))) ( -37.30) >pf5.74 6950066 112 + 7074893 AAUUUGGGUAUGUGAUAGAAAGAU-AGACUGAAC-GUUACUUUCACUGGUAACGGAUCAGGCUAAGGUAAAAUUUGUGAGUUCUCUUUUAAUUAAGAA--UGCAAAUUUUCGGCGA ((((((.((...((((.(((((((-((.((((.(-((((((......)))))))..)))).)))......((((....)))).)))))).))))...)--).))))))........ ( -23.10) >pfo.66 6314935 111 + 6438405 AAUUUGGGUAUGUGAUAGAAAGAU-AGACUGAAC-GUUACUUUCACUGGUAACGGAUCAGGCUAAGGUAAAAUUUGUGAGUUCUCUU--AAUUGAGAA-AUUCGAAUUUUCGGCGA ..........(((..........(-((.((((.(-((((((......)))))))..)))).)))..(.((((((((....(((((..--....)))))-...))))))))).))). ( -25.90) >consensus AAUUCGGGUAUGUGAUAGAA__AU_AGACUGAAC_AUCACUUUCACUGGUGAUGGAUCAGGCUAAGGUAAAAUUUGUGAGUUCU__U__AA__A__AA_GUGCGAAUUUUCGGCGA ............................((((.(.((((((......)))))).).))))(((...(.((((((((((.....((((......))))...)))))))))))))).. (-19.27 = -16.78 + -2.49)
| Location | 4,791,866 – 4,791,956 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 67.27 |
| Mean single sequence MFE | -25.58 |
| Consensus MFE | -14.17 |
| Energy contribution | -12.98 |
| Covariance contribution | -1.19 |
| Combinations/Pair | 1.40 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.55 |
| SVM decision value | 2.78 |
| SVM RNA-class probability | 0.996967 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4791866 90 - 6264404 CACCAUCU--AAAACAAUCGUCG--------AAAGCUCAGAAAUGAAUGUUC------------------GUGAAUGAACAUUGAUUUCUGGUC-UUUGCACCAGAACUGUUCUUUAAA ........--.........((((--------((..(.(((((((.(((((((------------------(....)))))))).))))))))..-)))).))................. ( -21.80) >pss.3053 5951255 108 - 6093698 CACCACUU--ACUGCUUCUGUUU------UGAAAGCUUAGAAAUGAGCAUUCCAUUGAUCCUGAUGAUCAGUGCAUGAAUGUUGAUUUCUAGUC-UUUG-AUUAGAUC-GUUCUUUAAA ........--......((((...------(.((((.((((((((.(((((((((((((((.....))))))))...))))))).)))))))).)-))).-).))))..-.......... ( -29.70) >pel.2704 5001798 97 - 5888780 CACCACUCUUUCAGGUUUCGCAGCACUGCUCAGAACUUAGAAAUGAGCAUUCGGUGA------------------UGAAUGUUGAUUUCUGACU-UUUG--UCAGAUC-GUUCUUUAAA ((((.(((((((((((((.(((....)))...)))))).)))).))).....)))).------------------.(((((......((((((.-...)--))))).)-))))...... ( -25.70) >pf5.74 6950178 85 + 7074893 CACCAUAA--ACUGCUUCUGAA-----------AGCUUAGAAAUGAGCAUUC-----------AC--------CUUGAAUGUUGAUUUCUAGUCUUUUG-AUUAGAUC-GUUCUUUAAA .....(((--(..((.((((((-----------((.((((((((.(((((((-----------..--------...))))))).)))))))).)))...-.)))))..-))..)))).. ( -21.70) >pfo.66 6315046 97 + 6438405 CACCACUA---CUGCUUCUGAAG------UAAAAGCUUAGAAAUGAGCAUUCCAUCG---UUGAU--------GGUGAAUGUUGAUUUCUAGUCUUUUG-ACUGGUUC-GUUCUUUAAA .((((..(---((........))------)(((((.((((((((.(((((((((((.---..)))--------))..)))))).)))))))).))))).-..))))..-.......... ( -24.40) >psp.2973 5782417 107 - 5928787 CACCACUU--ACUGCUUCUG-UU------UGAAAGCUUAGAAAUGAGCAUUCCAUUGAUCCUGAUGAUCAGUGCAUGAAUGUUGAUUUCUAGUC-UUUG-AUUAGAUC-GUUCUUUAAA ........--......((((-.(------..((..((.((((((.(((((((((((((((.....))))))))...))))))).))))))))..-))..-).))))..-.......... ( -30.20) >consensus CACCACUU__ACUGCUUCUGAAG______U_AAAGCUUAGAAAUGAGCAUUCCAU_G______AU________CAUGAAUGUUGAUUUCUAGUC_UUUG_AUUAGAUC_GUUCUUUAAA ....................................((((((((.(((((((........................))))))).))))))))........................... (-14.17 = -12.98 + -1.19)

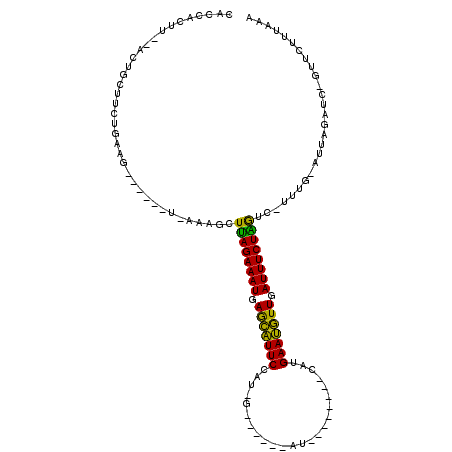
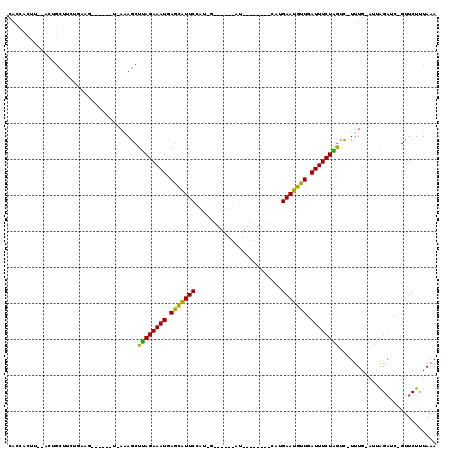
| Location | 4,791,905 – 4,791,996 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 71.60 |
| Mean single sequence MFE | -29.42 |
| Consensus MFE | -21.00 |
| Energy contribution | -20.53 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.895356 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4791905 91 - 6264404 UUUGCACGCAGGAGGUCAGGAGUUCGAUCCUCCUUGGCUCCACCAUCU--AAAACAAUCGUCG--------AAAGCUCAGAAAUGAAUGUUC------------------GUGAAUGAA ..........((((.(.(((((.......))))).).)))).......--....((.(((.((--------((...(((....)))...)))------------------)))).)).. ( -23.90) >pss.3053 5951292 111 - 6093698 CUUGCACGCAGGAGGUCAGCGGUUCGAUCCCGCUUGGCUCCACCACUU--ACUGCUUCUGUUU------UGAAAGCUUAGAAAUGAGCAUUCCAUUGAUCCUGAUGAUCAGUGCAUGAA ..((((((((((((...(((((.......)))))(((.....)))...--....)))))))..------.(((.(((((....))))).)))...(((((.....)))))))))).... ( -34.70) >pel.2704 5001834 101 - 5888780 CUUGCACGCAGGAGGUCAGCGGUUCGAUCCCGCUUGGCUCCACCACUCUUUCAGGUUUCGCAGCACUGCUCAGAACUUAGAAAUGAGCAUUCGGUGA------------------UGAA ....(((((.((((.(.(((((.......))))).).))))....(((((((((((((.(((....)))...)))))).)))).)))......))).------------------)).. ( -29.10) >pf5.74 6950216 87 + 7074893 CUUGCACGCAGGAGGUCAGCGGUUCGAUCCCGCUUGGCUCCACCAUAA--ACUGCUUCUGAA-----------AGCUUAGAAAUGAGCAUUC-----------AC--------CUUGAA ..........((((.(.(((((.......))))).).)))).......--........((((-----------.(((((....))))).)))-----------).--------...... ( -25.00) >pfo.66 6315084 99 + 6438405 CUUGCACGCAGGAGGUCAGCGGUUCGAUCCCGCUUGGCUCCACCACUA---CUGCUUCUGAAG------UAAAAGCUUAGAAAUGAGCAUUCCAUCG---UUGAU--------GGUGAA ((((....)))).(((((((((.......)))).))))).(((((.((---((........))------))...(((((....))))).........---....)--------)))).. ( -29.10) >psp.2973 5782454 110 - 5928787 CUUGCACGCAGGAGGUCAGCGGUUCGAUCCCGCUUGGCUCCACCACUU--ACUGCUUCUG-UU------UGAAAGCUUAGAAAUGAGCAUUCCAUUGAUCCUGAUGAUCAGUGCAUGAA ..((((((((((((...(((((.......)))))(((.....)))...--....))))))-).------.(((.(((((....))))).)))...(((((.....)))))))))).... ( -34.70) >consensus CUUGCACGCAGGAGGUCAGCGGUUCGAUCCCGCUUGGCUCCACCACUU__ACUGCUUCUGAAG______U_AAAGCUUAGAAAUGAGCAUUCCAU_G______AU________CAUGAA ..........((((.(.(((((.......))))).).)))).................................(((((....)))))............................... (-21.00 = -20.53 + -0.47)

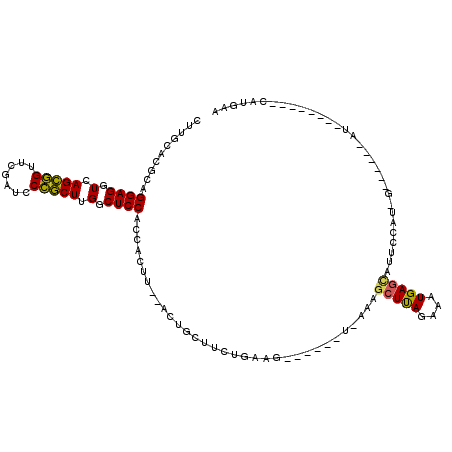
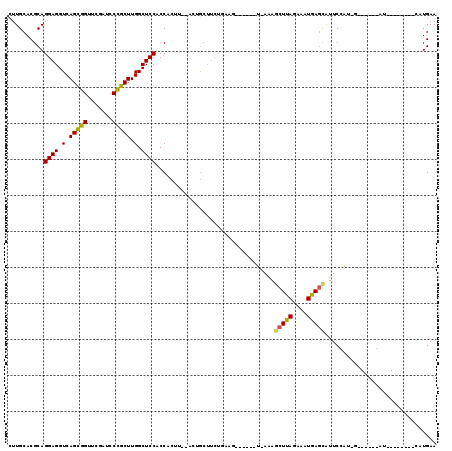
Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:09:30 2007