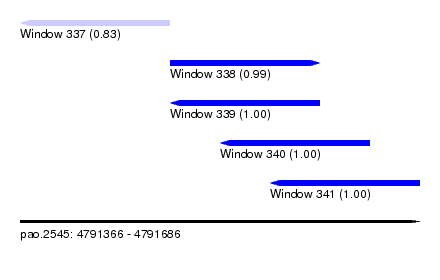
| Sequence ID | pao.2545 |
|---|---|
| Location | 4,791,366 – 4,791,686 |
| Length | 320 |
| Max. P | 0.999999 |

| Location | 4,791,366 – 4,791,486 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.78 |
| Mean single sequence MFE | -38.07 |
| Consensus MFE | -36.67 |
| Energy contribution | -33.02 |
| Covariance contribution | -3.66 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.828477 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4791366 120 - 6264404 UAGUAGUGGCGAGCGAACGGGGAUUAGCCCUUAAGCUUCAUUGAUUUUAGCGGAACGCUCUGGAAAGUGCGGCCAUAGUGGGUGAUAGCCCCGUACGCGAAAGGAUCUUUGAAGUGAAAU ............(((.((((((....((((....(((...........)))((..(((.((....)).))).)).....)))).....)))))).)))...................... ( -36.10) >pau.414 5802197 120 + 6537648 UAGUAGUGGCGAGCGAACGGGGAUUAGCCCUUAAGCUUCAUUGAUUUUAGCGGAACGCUCUGGAAAGUGCGGCCAUAGUGGGUGAUAGCCCCGUACGCGAAAGGAUCUUUGAAGUGAAAU ............(((.((((((....((((....(((...........)))((..(((.((....)).))).)).....)))).....)))))).)))...................... ( -36.10) >pss.3053 5950756 120 - 6093698 UAGUAGUGGCGAGCGAACGGGGACCAGCCCUUAAGUUGUAUUGAGAUUAGCGGAACGUUCUGGAAAGGACGGCCAUAGUGGGUGAUAGCCCUGUACGCGAAAAUCCCUUUGCAAUGAAAU ..((((.((.(((((.((((((....((((....((((.........))))((..((((((....)))))).)).....)))).....)))))).))).....)))).))))........ ( -38.00) >pel.2704 5001286 120 - 5888780 UAGUAGUGGCGAGCGAACGGGGACCAGCCCUUAAGCUGGUUUGAGAUUAGUGGAACGCUCUGGAAAGUGCGGCCAUAGUGGGUGAUAGCCCCGUACACGAAAAUCUCUUGCCAGUGAAAU ...((.((((((........((((((((......))))))))((((((.((((..(((.((....)).)))..)...(.((((....)))))...)))...)))))))))))).)).... ( -43.30) >ppk.338 5481656 120 + 6181863 UAGUAGUGGCGAGCGAACGGGGACCAGCCCUUAAGUUGAUUUGAGAUUAGUGGAACGCUCUGGAAAGUGCGGCCAUAGUGGGUGAUAGCCCCGUACACGAAAAUCUCUUGUCAAUGAAAU ......(((((((.((((((((....((((......(((((...)))))((((..(((.((....)).))).))))...)))).....)))))).........)).)))))))....... ( -36.90) >psp.2973 5781921 120 - 5928787 UAGUAGUGGCGAGCGAACGGGGACCAGCCCUUAAGUUGUAUUGAGAUUAGCGGAACGUUCUGGAAAGGACGGCCAUAGUGGGUGAUAGCCCUGUACGCGAAAAUCCCUUUGCAAUGAAAU ..((((.((.(((((.((((((....((((....((((.........))))((..((((((....)))))).)).....)))).....)))))).))).....)))).))))........ ( -38.00) >consensus UAGUAGUGGCGAGCGAACGGGGACCAGCCCUUAAGCUGCAUUGAGAUUAGCGGAACGCUCUGGAAAGUGCGGCCAUAGUGGGUGAUAGCCCCGUACGCGAAAAUCUCUUUGCAAUGAAAU ..................((((.....))))...((((((..((((((.((((..(((.((....)).)))..).....((((....))))....)))...))))))..))))))..... (-36.67 = -33.02 + -3.66)

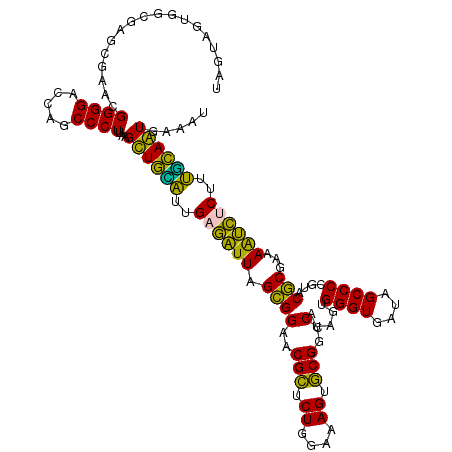
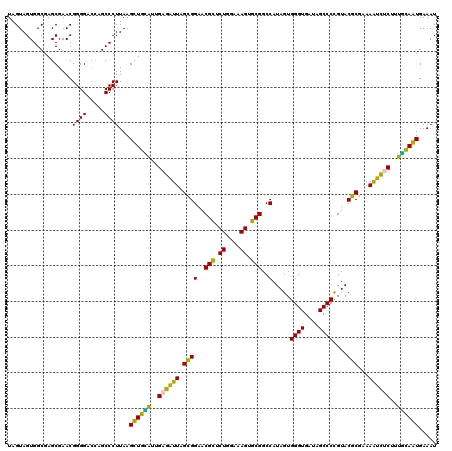
| Location | 4,791,486 – 4,791,606 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.06 |
| Mean single sequence MFE | -34.05 |
| Consensus MFE | -32.27 |
| Energy contribution | -30.67 |
| Covariance contribution | -1.61 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.40 |
| SVM RNA-class probability | 0.993415 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4791486 120 + 6264404 AGGGAAUCUCGGUUGAUUUCUUUUCCUCAGGGUACUUAGAUGUUUCAGUUCCCCUGGUUCGCCUCUUGCACCUAUGGAUUCAGUACAAGAUACCUAGGUUAUCCUAGGUGGGUUCCCCCA .((((((((..(((.....((..(((..((((((...((.((..((((.....))))..)).))..))).)))..)))...)).....)))(((((((....)))))))))))))))... ( -35.40) >pau.414 5802317 120 - 6537648 AGGGAAUCUCGGUUGAUUUCUUUUCCUCAGGGUACUUAGAUGUUUCAGUUCCCCUGGUUCGCCUCUUGCACCUAUGGAUUCAGUACAAGAUACCUAGGUUAUCCUAGGUGGGUUCCCCCA .((((((((..(((.....((..(((..((((((...((.((..((((.....))))..)).))..))).)))..)))...)).....)))(((((((....)))))))))))))))... ( -35.40) >pel.2704 5001406 120 + 5888780 AGGGAAUCUCGGUUGAUUUCUUUUCCUCAGGGUACUUAGAUGUUUCAGUUCCCCUGGUUCGCCUCUUGCACCUAUGUAUUCAGUACAAGAUACUCAGCUUGUGCUGAGUGGGUUCCCCCA .(((((((..(((.............((((((.(((..((....)))))..))))))...((.....)))))....(((((((((((((........))))))))))))))))))))... ( -38.80) >ppk.338 5481776 120 - 6181863 AGGGAAUCUCGGUUGAUUUCUUUUCCUCAGGGUACUUAGAUGUUUCAGUUCCCCUGGUUCGCCUCUUGCACCUAUGUAUUCAGUACAAGAUAACCAGCUUAUGCUGGCUGGGUUCCCCCA .(((((((.(((..............((((((.(((..((....)))))..))))))......(((((.((...........)).)))))...(((((....)))))))))))))))... ( -32.90) >pfo.66 6314655 120 - 6438405 AGGGAAUCUCGGUUGAUUUCUUUUCCUCAGGGUACUUAGAUGUUUCAGUUCCCCUGGUUCGCCUCUUGCACCUAUGUAUUCAGUACAAGAUAACCAUCUUAUGAUGGCUGGGUUCCCCCA .(((((((.(((..............((((((.(((..((....)))))..))))))......(((((.((...........)).)))))...(((((....)))))))))))))))... ( -30.90) >psp.2973 5782041 120 + 5928787 AGGGAAUCUCGGUUGAUUUCUUUUCCUCAGGGUACUUAGAUGUUUCAGUUCCCCUGGUUCGCCUCUUGCACCUAUGUAUUCAGUACAAGAUAACCAUCUUAUGAUGGCUGGGUUCCCCCA .(((((((.(((..............((((((.(((..((....)))))..))))))......(((((.((...........)).)))))...(((((....)))))))))))))))... ( -30.90) >consensus AGGGAAUCUCGGUUGAUUUCUUUUCCUCAGGGUACUUAGAUGUUUCAGUUCCCCUGGUUCGCCUCUUGCACCUAUGUAUUCAGUACAAGAUAACCAGCUUAUGCUGGCUGGGUUCCCCCA .((((((((.(((.(((..........(((((.(((..((....)))))..)))))))).)))(((((.((...........)).)))))...(((((....)))))..))))))))... (-32.27 = -30.67 + -1.61)

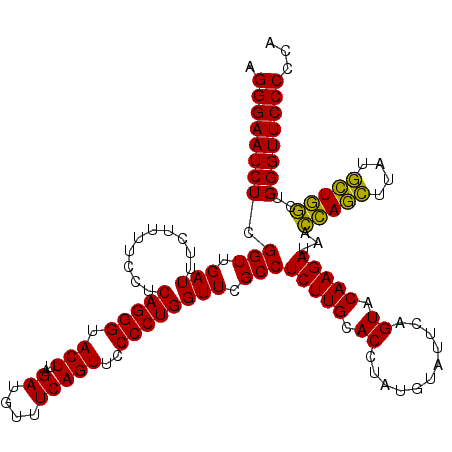
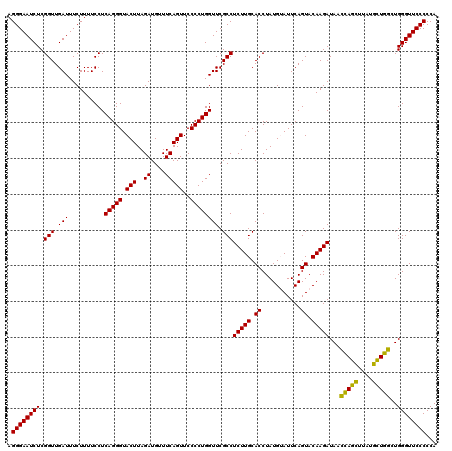
| Location | 4,791,486 – 4,791,606 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.06 |
| Mean single sequence MFE | -39.45 |
| Consensus MFE | -41.56 |
| Energy contribution | -39.45 |
| Covariance contribution | -2.11 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.43 |
| Structure conservation index | 1.05 |
| SVM decision value | 6.72 |
| SVM RNA-class probability | 0.999999 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4791486 120 - 6264404 UGGGGGAACCCACCUAGGAUAACCUAGGUAUCUUGUACUGAAUCCAUAGGUGCAAGAGGCGAACCAGGGGAACUGAAACAUCUAAGUACCCUGAGGAAAAGAAAUCAACCGAGAUUCCCU .(((....)))(((((((....))))))).(((((((((.........)))))))))((.(((.(((((..(((..........))).))))).((...(....)...))....))))). ( -40.00) >pau.414 5802317 120 + 6537648 UGGGGGAACCCACCUAGGAUAACCUAGGUAUCUUGUACUGAAUCCAUAGGUGCAAGAGGCGAACCAGGGGAACUGAAACAUCUAAGUACCCUGAGGAAAAGAAAUCAACCGAGAUUCCCU .(((....)))(((((((....))))))).(((((((((.........)))))))))((.(((.(((((..(((..........))).))))).((...(....)...))....))))). ( -40.00) >pel.2704 5001406 120 - 5888780 UGGGGGAACCCACUCAGCACAAGCUGAGUAUCUUGUACUGAAUACAUAGGUGCAAGAGGCGAACCAGGGGAACUGAAACAUCUAAGUACCCUGAGGAAAAGAAAUCAACCGAGAUUCCCU .(((....)))(((((((....))))))).(((((((((.........)))))))))((.(((.(((((..(((..........))).))))).((...(....)...))....))))). ( -40.40) >ppk.338 5481776 120 + 6181863 UGGGGGAACCCAGCCAGCAUAAGCUGGUUAUCUUGUACUGAAUACAUAGGUGCAAGAGGCGAACCAGGGGAACUGAAACAUCUAAGUACCCUGAGGAAAAGAAAUCAACCGAGAUUCCCU .(((....)))(((((((....))))))).(((((((((.........)))))))))((.(((.(((((..(((..........))).))))).((...(....)...))....))))). ( -40.10) >pfo.66 6314655 120 + 6438405 UGGGGGAACCCAGCCAUCAUAAGAUGGUUAUCUUGUACUGAAUACAUAGGUGCAAGAGGCGAACCAGGGGAACUGAAACAUCUAAGUACCCUGAGGAAAAGAAAUCAACCGAGAUUCCCU .(((....)))(((((((....))))))).(((((((((.........)))))))))((.(((.(((((..(((..........))).))))).((...(....)...))....))))). ( -38.10) >psp.2973 5782041 120 - 5928787 UGGGGGAACCCAGCCAUCAUAAGAUGGUUAUCUUGUACUGAAUACAUAGGUGCAAGAGGCGAACCAGGGGAACUGAAACAUCUAAGUACCCUGAGGAAAAGAAAUCAACCGAGAUUCCCU .(((....)))(((((((....))))))).(((((((((.........)))))))))((.(((.(((((..(((..........))).))))).((...(....)...))....))))). ( -38.10) >consensus UGGGGGAACCCACCCAGCAUAAGCUGGGUAUCUUGUACUGAAUACAUAGGUGCAAGAGGCGAACCAGGGGAACUGAAACAUCUAAGUACCCUGAGGAAAAGAAAUCAACCGAGAUUCCCU .(((....)))(((((((....))))))).(((((((((.........)))))))))((.(((.(((((..(((..........))).))))).((...(....)...))....))))). (-41.56 = -39.45 + -2.11)

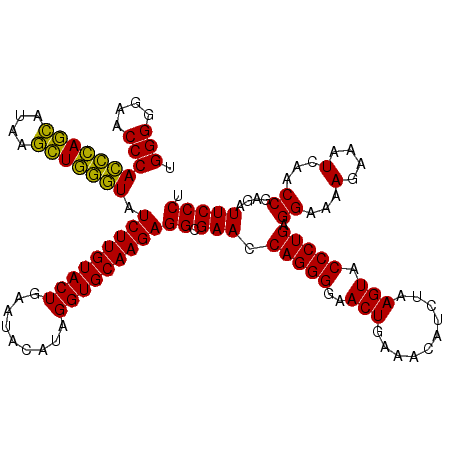
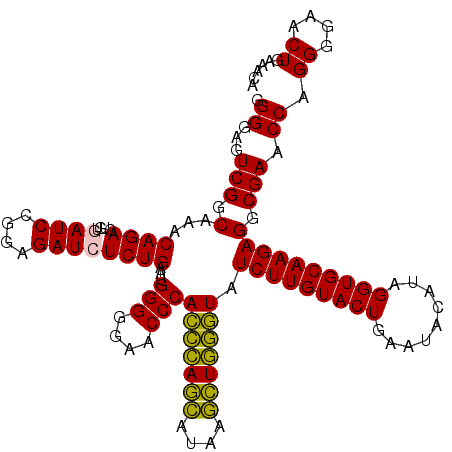
| Location | 4,791,526 – 4,791,646 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.67 |
| Mean single sequence MFE | -40.24 |
| Consensus MFE | -40.96 |
| Energy contribution | -39.18 |
| Covariance contribution | -1.77 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.69 |
| Structure conservation index | 1.02 |
| SVM decision value | 5.72 |
| SVM RNA-class probability | 0.999993 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4791526 120 - 6264404 GGGGAGUCGGCAAACAGACUUUGAUCCGGAGAUCUCUGAAUGGGGGAACCCACCUAGGAUAACCUAGGUAUCUUGUACUGAAUCCAUAGGUGCAAGAGGCGAACCAGGGGAACUGAAACA .((...(((.(...((((....((((....))))))))...(((....)))(((((((....))))))).(((((((((.........)))))))))).))).)).((....))...... ( -40.70) >pau.414 5802357 120 + 6537648 GGGGAGUCGGCAAACAGACUUUGAUCCGGAGAUCUCUGAAUGGGGGAACCCACCUAGGAUAACCUAGGUAUCUUGUACUGAAUCCAUAGGUGCAAGAGGCGAACCAGGGGAACUGAAACA .((...(((.(...((((....((((....))))))))...(((....)))(((((((....))))))).(((((((((.........)))))))))).))).)).((....))...... ( -40.70) >pel.2704 5001446 120 - 5888780 GGGGAGUCGGCAAACAGACUGUGAUCCAGAGAUCUCUGAAUGGGGGAACCCACUCAGCACAAGCUGAGUAUCUUGUACUGAAUACAUAGGUGCAAGAGGCGAACCAGGGGAACUGAAACA .((...(((.(...((((....((((....))))))))...(((....)))(((((((....))))))).(((((((((.........)))))))))).))).)).((....))...... ( -41.10) >ppk.338 5481816 120 + 6181863 GGGGAGUCGGCAAACAGACUGUGAUCCAGAGAUCUCUGAAUGGGGGAACCCAGCCAGCAUAAGCUGGUUAUCUUGUACUGAAUACAUAGGUGCAAGAGGCGAACCAGGGGAACUGAAACA .((...(((.(...((((....((((....))))))))...(((....)))(((((((....))))))).(((((((((.........)))))))))).))).)).((....))...... ( -40.80) >pfo.66 6314695 120 + 6438405 GGGGAGUCGGCAAACAGACUUUGAUCCGGAGAUGUCUGAAUGGGGGAACCCAGCCAUCAUAAGAUGGUUAUCUUGUACUGAAUACAUAGGUGCAAGAGGCGAACCAGGGGAACUGAAACA .((...(((.(...(((((.((........)).)))))...(((....)))(((((((....))))))).(((((((((.........)))))))))).))).)).((....))...... ( -39.20) >psp.2973 5782081 120 - 5928787 GGGGAGUCGGCAAACAGACUGUGAUCCGGAGAUGUCUGAAUGGGGGAACCCAGCCAUCAUAAGAUGGUUAUCUUGUACUGAAUACAUAGGUGCAAGAGGCGAACCAGGGGAACUGAAACA .((...(((.(...(((((..............)))))...(((....)))(((((((....))))))).(((((((((.........)))))))))).))).)).((....))...... ( -38.94) >consensus GGGGAGUCGGCAAACAGACUGUGAUCCGGAGAUCUCUGAAUGGGGGAACCCACCCAGCAUAAGCUGGGUAUCUUGUACUGAAUACAUAGGUGCAAGAGGCGAACCAGGGGAACUGAAACA .((...(((.(...((((....((((....))))))))...(((....)))(((((((....))))))).(((((((((.........)))))))))).))).)).((....))...... (-40.96 = -39.18 + -1.77)

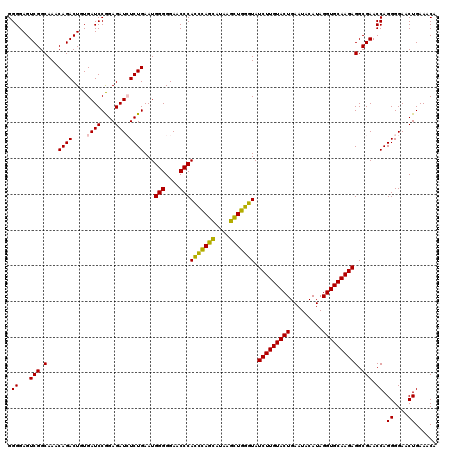
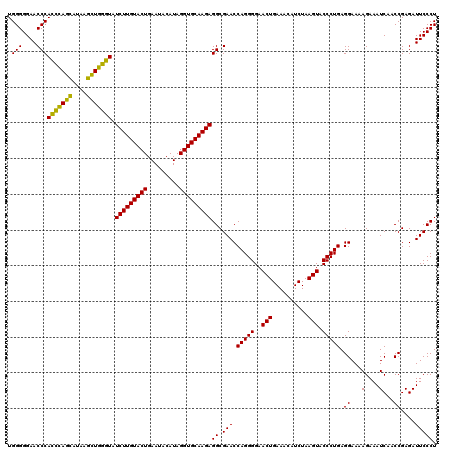
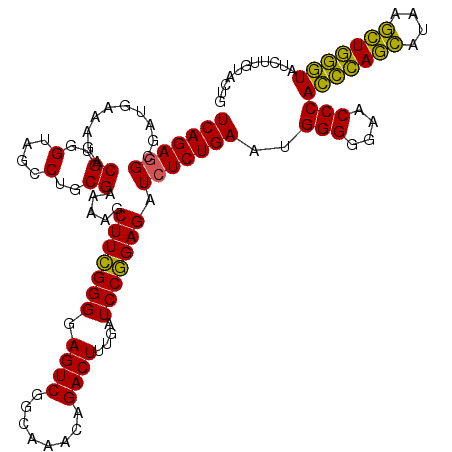
| Location | 4,791,566 – 4,791,686 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.39 |
| Mean single sequence MFE | -41.50 |
| Consensus MFE | -43.23 |
| Energy contribution | -40.85 |
| Covariance contribution | -2.38 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.00 |
| Structure conservation index | 1.04 |
| SVM decision value | 4.81 |
| SVM RNA-class probability | 0.999952 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4791566 120 - 6264404 UCAGAGGCGAUGAAAGACGUGGUAGCCUGCGAAAAGCUUCGGGGAGUCGGCAAACAGACUUUGAUCCGGAGAUCUCUGAAUGGGGGAACCCACCUAGGAUAACCUAGGUAUCUUGUACUG (((((((..........((..(....)..)).....((((((((((((........)))))...))))))).)))))))..(((....)))(((((((....)))))))........... ( -43.10) >pau.414 5802397 120 + 6537648 UCAGAGGCGAUGAAAGACGUGGUAGCCUGCGAAAAGCUUCGGGGAGUCGGCAAACAGACUUUGAUCCGGAGAUCUCUGAAUGGGGGAACCCACCUAGGAUAACCUAGGUAUCUUGUACUG (((((((..........((..(....)..)).....((((((((((((........)))))...))))))).)))))))..(((....)))(((((((....)))))))........... ( -43.10) >pel.2704 5001486 120 - 5888780 UCAGAGGCGAUGAAAGACGUGGUAGCCUGCGAUAAGCUUUGGGGAGUCGGCAAACAGACUGUGAUCCAGAGAUCUCUGAAUGGGGGAACCCACUCAGCACAAGCUGAGUAUCUUGUACUG (((((((..........((..(....)..)).....(((((((.((((........))))....))))))).)))))))..(((....)))(((((((....)))))))........... ( -41.60) >ppk.338 5481856 120 + 6181863 UCAGAGGCGAUGAAAGACGUGGUAGCCUGCGAUAAGCUUUGGGGAGUCGGCAAACAGACUGUGAUCCAGAGAUCUCUGAAUGGGGGAACCCAGCCAGCAUAAGCUGGUUAUCUUGUACUG (((((((..........((..(....)..)).....(((((((.((((........))))....))))))).)))))))..(((....)))(((((((....)))))))........... ( -41.30) >pf5.74 6949866 120 + 7074893 UCAGAGGCGAUGAAAGACGUGGUAGCCUGCGAUAAGCUUCGGGGAGUCGGCAAACAGACUUUGAUCCGGAGAUCUCUGAAUGGGGGAACCCAGCCAUCAUAAGAUGGUUAUCUUGUACUG (((((((..........((..(....)..)).....((((((((((((........)))))...))))))).)))))))..(((....)))(((((((....)))))))........... ( -41.20) >pfo.66 6314735 120 + 6438405 UCAGAGGCGAUGAAAGACGUGGUAGCCUGCGAAAAGCUUCGGGGAGUCGGCAAACAGACUUUGAUCCGGAGAUGUCUGAAUGGGGGAACCCAGCCAUCAUAAGAUGGUUAUCUUGUACUG .(((.(((......(((((..(....)..)......((((((((((((........)))))...)))))))..))))...((((....)))))))...(((((((....))))))).))) ( -38.70) >consensus UCAGAGGCGAUGAAAGACGUGGUAGCCUGCGAAAAGCUUCGGGGAGUCGGCAAACAGACUUUGAUCCGGAGAUCUCUGAAUGGGGGAACCCACCCAGCAUAAGCUGGGUAUCUUGUACUG (((((((..........((..(....)..)).....(((((((.((((........))))....))))))).)))))))..(((....)))(((((((....)))))))........... (-43.23 = -40.85 + -2.38)

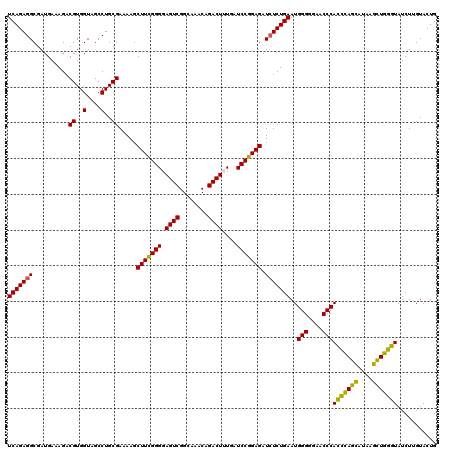
Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:09:27 2007