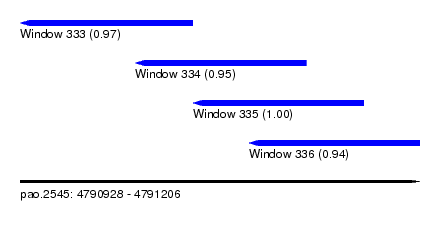
| Sequence ID | pao.2545 |
|---|---|
| Location | 4,790,928 – 4,791,206 |
| Length | 278 |
| Max. P | 0.999941 |

| Location | 4,790,928 – 4,791,048 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.44 |
| Mean single sequence MFE | -36.17 |
| Consensus MFE | -34.97 |
| Energy contribution | -33.97 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968534 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
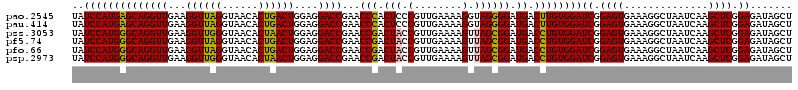
>pao.2545 4790928 120 - 6264404 UAUCCAUGAGCAGGUUGAAGGUUAGGUAACACUGACUGGAGGACCGAACCCACUCCCGUUGAAAAGGUAGGGGAUGACUUGUGGAUCGGAGUGAAAGGCUAAUCAAGCUCGGAGAUAGCU ..((((..((((((((...((((((......))))))....))))...(((.((.((........)).))))).)).))..))))((.((((..............)))).))....... ( -35.84) >pau.414 5801759 120 + 6537648 UAUCCAUGAGCAGGUUGAAGGUUAGGUAACACUGACUGGAGGACCGAACCCACUCCCGUUGAAAAGGUAGGGGAUGACUUGUGGAUCGGAGUGAAAGGCUAAUCAAGCUCGGAGAUAGCU ..((((..((((((((...((((((......))))))....))))...(((.((.((........)).))))).)).))..))))((.((((..............)))).))....... ( -35.84) >pss.3053 5950317 120 - 6093698 UAUCCAUGGGCAGGUUGAAGGUUGGGUAACACUAACUGGAGGACCGAACCGACUACCGUUGAAAAGUUAGCGGAUGACCUGUGGAUCGGAGUGAAAGGCUAAUCAAGCUCGGAGAUAGCU ..((((((((((((((...((((((......))))))....))))..........(((((((....))))))).)).))))))))((.((((..............)))).))....... ( -36.14) >pf5.74 6949227 120 + 7074893 UAUCCAUGGGCAGGUUGAAGGUUAGGUAACACUGACUGGAGGACCGAACCGACUACCGUUGAAAAGUUAGCGGAUGACCUGUGGAUCGGAGUGAAAGGCUAAUCAAGCUCGGAGAUAGCU ..((((((((((((((...((((((......))))))....))))..........(((((((....))))))).)).))))))))((.((((..............)))).))....... ( -36.54) >pfo.66 6314096 120 + 6438405 UAUCCAUGGGCAGGUUGAAGGUUAGGUAACACUGACUGGAGGACCGAACCGACUACCGUUGAAAAGUUAGCGGAUGACCUGUGGAUCGGAGUGAAAGGCUAAUCAAGCUCGGAGAUAGCU ..((((((((((((((...((((((......))))))....))))..........(((((((....))))))).)).))))))))((.((((..............)))).))....... ( -36.54) >psp.2973 5781482 120 - 5928787 UAUCCAUGGGCAGGUUGAAGGUUGGGUAACACUAACUGGAGGACCGAACCGACUACCGUUGAAAAGUUAGCGGAUGACCUGUGGAUCGGAGUGAAAGGCUAAUCAAGCUCGGAGAUAGCU ..((((((((((((((...((((((......))))))....))))..........(((((((....))))))).)).))))))))((.((((..............)))).))....... ( -36.14) >consensus UAUCCAUGGGCAGGUUGAAGGUUAGGUAACACUGACUGGAGGACCGAACCGACUACCGUUGAAAAGUUAGCGGAUGACCUGUGGAUCGGAGUGAAAGGCUAAUCAAGCUCGGAGAUAGCU ..((((((((((((((...((((((......))))))....))))...(((.(((.(........).)))))).)).))))))))((.((((..............)))).))....... (-34.97 = -33.97 + -1.00)
| Location | 4,791,008 – 4,791,127 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.59 |
| Mean single sequence MFE | -38.45 |
| Consensus MFE | -36.20 |
| Energy contribution | -35.87 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.949115 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4791008 119 - 6264404 ACCGUAUAGGGUAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCG-UUUAGUCGCUGGGUAUAGACCCGAAACCGGGCGAUCUAUCCAUGAGCAGGUUGAAGGUUAGGUAACACUGACUGGA .((((.((((((((((.((((((......((((.....)))).-.....)))))).))...(.((((....)))))...)))))).)).))........((((((......)))))))). ( -38.40) >pau.414 5801839 119 + 6537648 ACCGUAUAGGGUAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCG-UUUAGUCGCUGGGUAUAGACCCGAAACCGGGCGAUCUAUCCAUGAGCAGGUUGAAGGUUAGGUAACACUGACUGGA .((((.((((((((((.((((((......((((.....)))).-.....)))))).))...(.((((....)))))...)))))).)).))........((((((......)))))))). ( -38.40) >pss.3053 5950397 119 - 6093698 ACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCG-UCUAGUCGCUGGGAAUAGACCCGAAACCGGGCGAUCUAUCCAUGGGCAGGUUGAAGGUUGGGUAACACUAACUGGA .(((....((((((((...((....))..))))).....((((-......)))).........))).....))).((((((..(....)..))))))..((((((......))))))... ( -37.70) >pel.2704 5000926 120 - 5888780 ACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCGCUUUAGUCGCUGGGUAUAGACCCGAAACCGGGCGAUCUAUCCAUGGGCAGGUUGAAGGUUAGGUAACACUGACUGGA .(((....(((...((.(((((((((((..(((....))).)))))...)))))).)).....))).....))).((((((..(....)..))))))..((((((......))))))... ( -40.20) >ppk.338 5481297 119 + 6181863 ACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCG-UUUAGUCGCUGGGUAUAGACCCGAAACCGGGCGAUCUAUCCAUGGGCAGGUUGAAGGUUAGGUAACACUGACUGGA .(((....(((...((.((((((......((((.....)))).-.....)))))).)).....))).....))).((((((..(....)..))))))..((((((......))))))... ( -38.30) >psp.2973 5781562 119 - 5928787 ACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCG-UCUAGUCGCUGGGAAUAGACCCGAAACCGGGCGAUCUAUCCAUGGGCAGGUUGAAGGUUGGGUAACACUAACUGGA .(((....((((((((...((....))..))))).....((((-......)))).........))).....))).((((((..(....)..))))))..((((((......))))))... ( -37.70) >consensus ACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCG_UUUAGUCGCUGGGUAUAGACCCGAAACCGGGCGAUCUAUCCAUGGGCAGGUUGAAGGUUAGGUAACACUGACUGGA (((........(((((...((....))..))))).....(((......)))(((.((.(((((((((....))))...)))))))...))).)))....((((((......))))))... (-36.20 = -35.87 + -0.33)

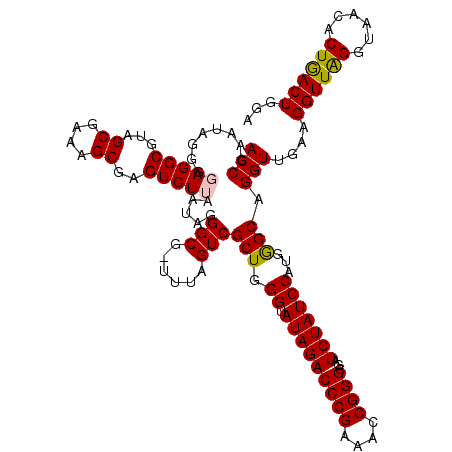
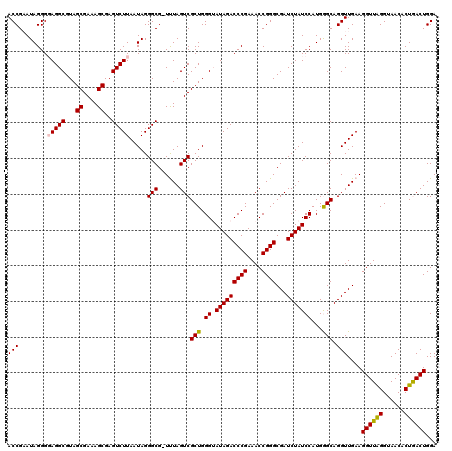
| Location | 4,791,048 – 4,791,167 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.98 |
| Mean single sequence MFE | -42.22 |
| Consensus MFE | -41.71 |
| Energy contribution | -41.27 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.99 |
| SVM decision value | 4.71 |
| SVM RNA-class probability | 0.999941 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4791048 119 - 6264404 UGUAUAAUGGGUCAGCGACUUAUAUUCAGUGGCAAGCUUAACCGUAUAGGGUAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCG-UUUAGUCGCUGGGUAUAGACCCGAAACCGGGCGAUC .........((((.......(((((((((((((..(((((.((.....)).)))))...((....))..((((.....)))).-....)))))))))))))(.((((....))))))))) ( -41.70) >pau.414 5801879 119 + 6537648 UGUAUAAUGGGUCAGCGACUUAUAUUCAGUGGCAAGCUUAACCGUAUAGGGUAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCG-UUUAGUCGCUGGGUAUAGACCCGAAACCGGGCGAUC .........((((.......(((((((((((((..(((((.((.....)).)))))...((....))..((((.....)))).-....)))))))))))))(.((((....))))))))) ( -41.70) >pel.2704 5000966 120 - 5888780 UGUAUAAUGGGUCAGCGACUUAUAUUCAGUGGCGAGCUUAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCGCUUUAGUCGCUGGGUAUAGACCCGAAACCGGGCGAUC .........((((.......(((((((((((((..((((..((.....))..))))((.((....))..((((.....))))))....)))))))))))))(.((((....))))))))) ( -42.70) >ppk.338 5481337 119 + 6181863 UGUAUAAUGGGUCAGCGACUUAUAUUCAGUGGCGAGCUUAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCG-UUUAGUCGCUGGGUAUAGACCCGAAACCGGGCGAUC .........((((.......(((((((((((((..((((..((.....))..))))...((....))..((((.....)))).-....)))))))))))))(.((((....))))))))) ( -42.40) >pf5.74 6949347 119 + 7074893 UGUAUAAUGGGUCAGCGACUUAUUUUCAGUGGCAAGCUUAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCG-UCUAGUCGCUGGGAAUAGACCCGAAACCGGGCGAUC .........((((.......((((..(((((((..((((..((.....))..))))...((....))((((((.....)))).-))..)))))))..))))(.((((....))))))))) ( -42.40) >psp.2973 5781602 119 - 5928787 UGUAUAAUGGGUCAGCGACUUAUUUUCAGUGGCGAGCUUAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCG-UCUAGUCGCUGGGAAUAGACCCGAAACCGGGCGAUC .........((((.......((((..(((((((..((((..((.....))..))))...((....))((((((.....)))).-))..)))))))..))))(.((((....))))))))) ( -42.40) >consensus UGUAUAAUGGGUCAGCGACUUAUAUUCAGUGGCAAGCUUAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCG_UUUAGUCGCUGGGUAUAGACCCGAAACCGGGCGAUC .........((((.......(((((((((((((..((((..((.....))..))))...((....))..((((.....))))......)))))))))))))(.((((....))))))))) (-41.71 = -41.27 + -0.44)

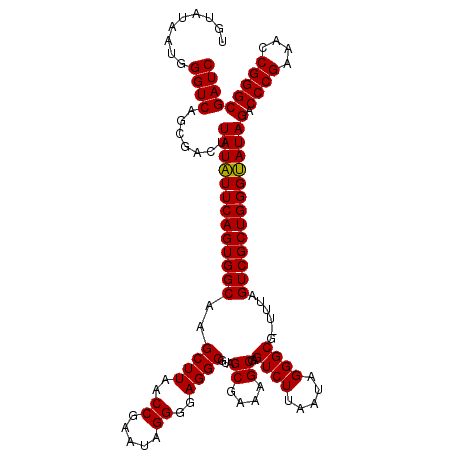
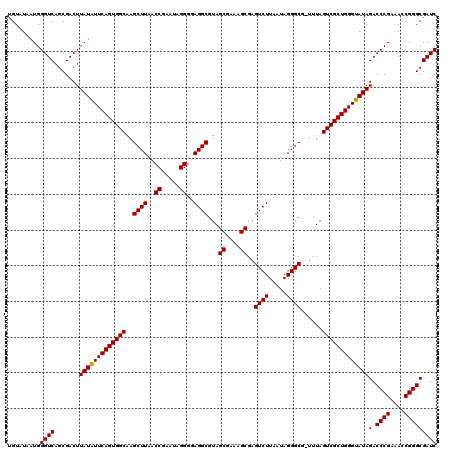
| Location | 4,791,087 – 4,791,206 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.50 |
| Mean single sequence MFE | -36.40 |
| Consensus MFE | -36.18 |
| Energy contribution | -35.07 |
| Covariance contribution | -1.11 |
| Combinations/Pair | 1.11 |
| Mean z-score | -0.99 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938717 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4791087 119 - 6264404 AGCAGUGGGAGCCUAC-UUGUUAGGUGACUGCGUACCUUUUGUAUAAUGGGUCAGCGACUUAUAUUCAGUGGCAAGCUUAACCGUAUAGGGUAGGCGUAGCGAAAGCGAGUCUUAAUAGG ((((((((....))))-.))))....((((((.(((((..(((((..(((((..((.(((.......))).))..)))))...)))))))))).))...((....)).))))........ ( -35.10) >pau.414 5801918 119 + 6537648 AGCAGUGGGAGCCUAC-UUGUUAGGUGACUGCGUACCUUUUGUAUAAUGGGUCAGCGACUUAUAUUCAGUGGCAAGCUUAACCGUAUAGGGUAGGCGUAGCGAAAGCGAGUCUUAAUAGG ((((((((....))))-.))))....((((((.(((((..(((((..(((((..((.(((.......))).))..)))))...)))))))))).))...((....)).))))........ ( -35.10) >pss.3053 5950476 120 - 6093698 AGCAGUGGGAGCCCACUUUGUUGGGUGACUGCGUACCUUUUGUAUAAUGGGUCAGCGACUUAUUUUCAGUGGCGAGCUUAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUAGG .((.(..(..(((((......)))))..)..)(.((((..........))))).))((((..(((((....((..((((..((.....))..))))...)))))))..))))........ ( -38.20) >pf5.74 6949386 120 + 7074893 AGCAGUGGGAGCAGACUUUGUUCUGUGACUGCGUACCUUUUGUAUAAUGGGUCAGCGACUUAUUUUCAGUGGCAAGCUUAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUAGG .((.(..(..(((((......)))))..)..)(.((((..........))))).))((((..(((((.((.....((((..((.....))..))))...)))))))..))))........ ( -35.70) >pfo.66 6314255 120 + 6438405 AGCAGUGGGAGCAGACUUUGUUCUGUGACUGCGUACCUUUUGUAUAAUGGGUCAGCGACUUAUUUUCAGUGGCGAGCUUAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUAGG .((.(..(..(((((......)))))..)..)(.((((..........))))).))((((..(((((....((..((((..((.....))..))))...)))))))..))))........ ( -36.10) >psp.2973 5781641 120 - 5928787 AGCAGUGGGAGCCCACUUUGUUGGGUGACUGCGUACCUUUUGUAUAAUGGGUCAGCGACUUAUUUUCAGUGGCGAGCUUAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUAGG .((.(..(..(((((......)))))..)..)(.((((..........))))).))((((..(((((....((..((((..((.....))..))))...)))))))..))))........ ( -38.20) >consensus AGCAGUGGGAGCCCACUUUGUUAGGUGACUGCGUACCUUUUGUAUAAUGGGUCAGCGACUUAUUUUCAGUGGCAAGCUUAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUAGG .((.(..(..(((((......)))))..)..)(.((((..........))))).))(((((..............((((..((.....))..))))...((....)))))))........ (-36.18 = -35.07 + -1.11)

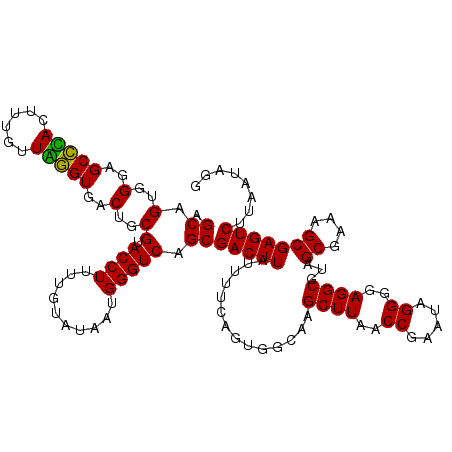
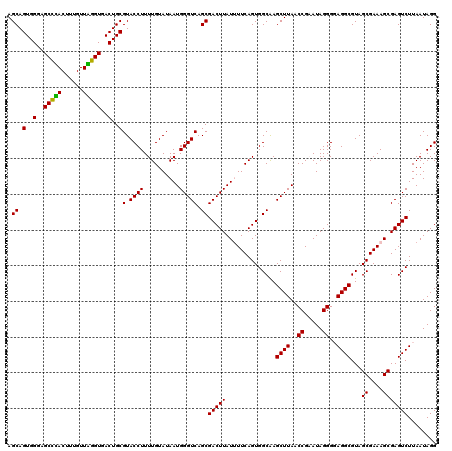
Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:09:22 2007