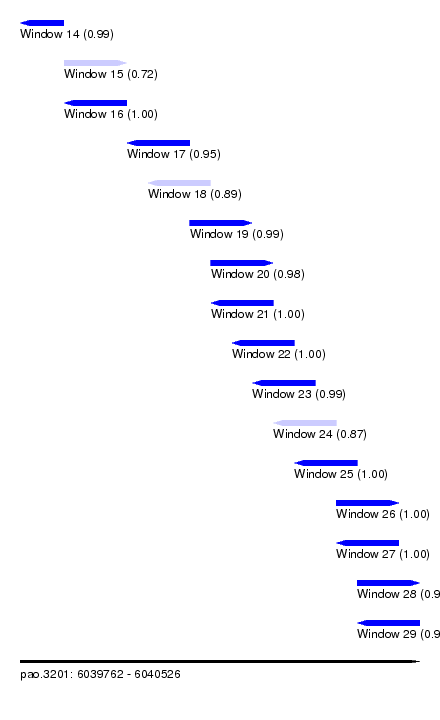
| Sequence ID | pao.3201 |
|---|---|
| Location | 6,039,762 – 6,040,526 |
| Length | 764 |
| Max. P | 0.999960 |

| Location | 6,039,762 – 6,039,846 |
|---|---|
| Length | 84 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 62.49 |
| Mean single sequence MFE | -27.82 |
| Consensus MFE | -11.93 |
| Energy contribution | -12.19 |
| Covariance contribution | 0.26 |
| Combinations/Pair | 1.48 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.43 |
| SVM decision value | 2.38 |
| SVM RNA-class probability | 0.993225 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
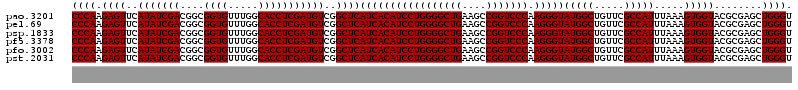
>pao.3201 6039762 84 - 6264404 AUAACACCCAAACAAUCUG---------------------ACGAUUGUGUGUUGUAAGGUGAAGUCG--ACGAACCGAAAGUUCG------CAUGAACC----GCAAAACACCUUGA (((((((....((((((..---------------------..)))))))))))))((((((......--..(((.(....))))(------(.......----))....)))))).. ( -22.70) >pss.1919 3847611 101 - 6093698 AUAACACCCAAGCAAUUUGCGUCGAAUG------------ACCAGAUUGCGGUG-ACUGUGAAGAUGACACGAACCGAAAGUUUG---CGUCACGAACGACACCUGAAACAGCUUGA ........(((((((((((.(((....)------------))))))))..((((-..(((...(.((((.((((.(....)))))---.)))))..))).)))).......))))). ( -29.30) >pel.69 5768297 81 + 5888780 AUAACACCCAAGCAAUCUGCACAUGAAC-------------GCAGAUUGUGGUG----GUGAAGACGAAA-GAACCGAAAGUUCGCAGCUUCACAAAGA------------------ ....((((...(((((((((........-------------)))))))))))))----((((((.(....-(((.(....))))...))))))).....------------------ ( -30.00) >psp.1833 3631614 103 - 5928787 AUAACACCCAAGCAAUUUGAGUGGACUUCCUGACAGGAGCAUCAGAUUGCGGUG-ACUGUGAAGACGACACGAACCGAAAGUUUG---CGUCACAAACG----------CACCGAGC ...........(((((((((...(.((((......))))).)))))))))((((-.((....))..(((.((((.(....)))))---.))).......----------)))).... ( -29.10) >pf5.3378 6382921 82 - 7074893 AUAACACCCAAGCAAUUUGCUUAAG--------------AAGCGAGUUGCGGUG---UGUGAAGACGAUACAAACCGAAAGUUUG--------CGAACA----------CACAAAUC ...(((((...((((((((((....--------------.))))))))))))))---)(((....((...((((.(....)))))--------))....----------)))..... ( -23.20) >pst.2031 3869612 101 - 6397126 AUAACACCCAAGCAAUUUGAGUCGAAUG------------ACCAGAUUGCGGUG-ACUGUGGAGAUGACACGAACCGAAGGUUUG---CGUCACGAACGACACCUGCAACAGCUUGC ........(((((((((((.(((....)------------))))))))((((((-..(((...(.((((.((((((...))))))---.)))))..))).)))).))....))))). ( -32.60) >consensus AUAACACCCAAGCAAUUUGCGUAGAAU_____________ACCAGAUUGCGGUG_ACUGUGAAGACGACACGAACCGAAAGUUUG___CGUCACGAACG__________CACCUUGC ....((((...((((((((.......................))))))))))))....((((........(((((.....))))).....))))....................... (-11.93 = -12.19 + 0.26)

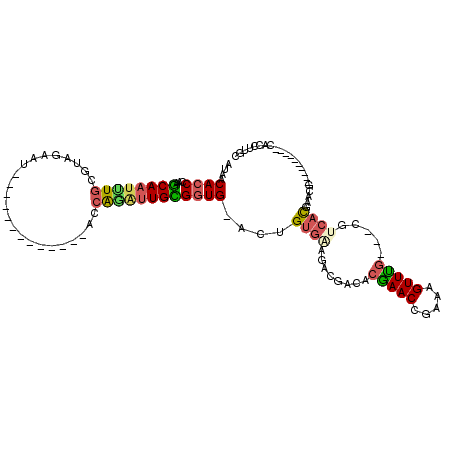
| Location | 6,039,846 – 6,039,966 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.44 |
| Mean single sequence MFE | -33.10 |
| Consensus MFE | -33.63 |
| Energy contribution | -32.64 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.10 |
| Mean z-score | -0.63 |
| Structure conservation index | 1.02 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.718021 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.3201 6039846 120 + 6264404 AUGGUCAAGCCUCACGGGCAAUUAGUACUGGUUAGCUCAACGCCUCACAACGCUUACACACCCAGCCUAUCAACGUCGUAGUCUUCGACGGCCCUUCAGGGGAAUCAAGUUCCCAGUGAG ..........(((((((((...(((..((((..(((...............))).......)))).)))....(((((.......))))))))).....((((((...)))))).))))) ( -32.36) >pau.3340 6312765 120 + 6537648 AUGGUCAAGCCUCACGGGCAAUUAGUACUGGUUAGCUCAACGCCUCACAACGCUUACACACCCAGCCUAUCAACGUCGUAGUCUUCGACGGCCCUUCAGGGGAAUCAAGUUCCCAGUGAG ..........(((((((((...(((..((((..(((...............))).......)))).)))....(((((.......))))))))).....((((((...)))))).))))) ( -32.36) >psp.1833 3631717 120 + 5928787 AUGGUCAAGCCUCACGGGCAAUUAGUAUUGGUUAGCUCAACGCCUCACAGCGCUUACACACCCAACCUAUCAACGUCGUAGUCUUCGACGGCCCUUUAGGGAACUCAAGGUUCCAGUGAG ..........(((((((((...(((..((((.........(((......))).........)))).)))....(((((.......))))))))).....(((((.....))))).))))) ( -33.47) >pf5.3378 6383003 120 + 7074893 AUGGUCAAGCCUCACGGGCAAUUAGUAUUGGUUAGCUCAACGCCUCACAGCGCUUACACACCCAACCUAUCAACGUCGUAGUCUUCGACGGCCCUUUAGGGAACUCAAGGUUCCAGUGAG ..........(((((((((...(((..((((.........(((......))).........)))).)))....(((((.......))))))))).....(((((.....))))).))))) ( -33.47) >pfo.3002 5726624 120 + 6438405 AUGGUCAAGCCUCACGGGCAAUUAGUAUUGGUUAGCUCAACGCCUCACAGCGCUUACACACCCAACCUAUCAACGUCGUAGUCUUCGACGGCCCUUCAGGGAACUCAAGGUUCCAGUGAG ..........(((((((((...(((..((((.........(((......))).........)))).)))....(((((.......))))))))).....(((((.....))))).))))) ( -33.47) >pst.2031 3869713 120 + 6397126 AUGGUCAAGCCUCACGGGCAAUUAGUAUUGGUUAGCUCAACGCCUCACAGCGCUUACACACCCAACCUAUCAACGUCGUAGUCUUCGACGGCCCUUCAGGGAACUCAAGGUUCCAGUGAG ..........(((((((((...(((..((((.........(((......))).........)))).)))....(((((.......))))))))).....(((((.....))))).))))) ( -33.47) >consensus AUGGUCAAGCCUCACGGGCAAUUAGUAUUGGUUAGCUCAACGCCUCACAGCGCUUACACACCCAACCUAUCAACGUCGUAGUCUUCGACGGCCCUUCAGGGAACUCAAGGUUCCAGUGAG ..........(((((((((...(((..((((.........(((......))).........)))).)))....(((((.......))))))))).....(((((.....))))).))))) (-33.63 = -32.64 + -1.00)

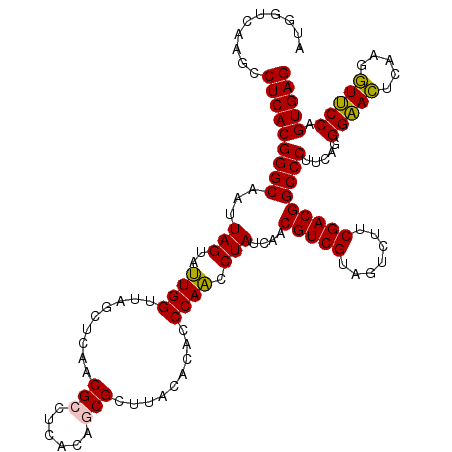
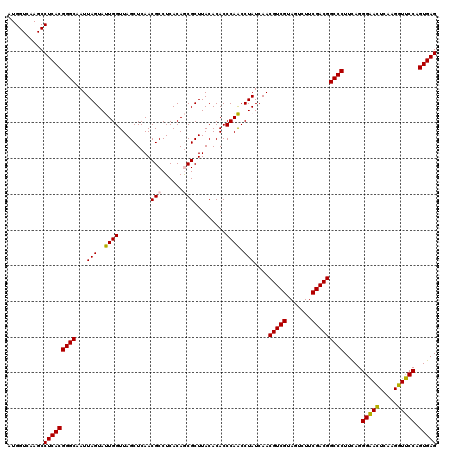
| Location | 6,039,846 – 6,039,966 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.44 |
| Mean single sequence MFE | -41.70 |
| Consensus MFE | -43.25 |
| Energy contribution | -41.70 |
| Covariance contribution | -1.55 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.20 |
| Structure conservation index | 1.04 |
| SVM decision value | 2.61 |
| SVM RNA-class probability | 0.995729 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.3201 6039846 120 - 6264404 CUCACUGGGAACUUGAUUCCCCUGAAGGGCCGUCGAAGACUACGACGUUGAUAGGCUGGGUGUGUAAGCGUUGUGAGGCGUUGAGCUAACCAGUACUAAUUGCCCGUGAGGCUUGACCAU (((((.(((((.....))))).....(((((((((.......)))))....((((((((..((...((((((....))))))..))...))))).)))...))))))))).......... ( -41.70) >pau.3340 6312765 120 - 6537648 CUCACUGGGAACUUGAUUCCCCUGAAGGGCCGUCGAAGACUACGACGUUGAUAGGCUGGGUGUGUAAGCGUUGUGAGGCGUUGAGCUAACCAGUACUAAUUGCCCGUGAGGCUUGACCAU (((((.(((((.....))))).....(((((((((.......)))))....((((((((..((...((((((....))))))..))...))))).)))...))))))))).......... ( -41.70) >psp.1833 3631717 120 - 5928787 CUCACUGGAACCUUGAGUUCCCUAAAGGGCCGUCGAAGACUACGACGUUGAUAGGUUGGGUGUGUAAGCGCUGUGAGGCGUUGAGCUAACCAAUACUAAUUGCCCGUGAGGCUUGACCAU (((((.(((((.....))))).....(((((((((.......)))))....((((((((..((...((((((....))))))..))...))))).)))...))))))))).......... ( -41.70) >pf5.3378 6383003 120 - 7074893 CUCACUGGAACCUUGAGUUCCCUAAAGGGCCGUCGAAGACUACGACGUUGAUAGGUUGGGUGUGUAAGCGCUGUGAGGCGUUGAGCUAACCAAUACUAAUUGCCCGUGAGGCUUGACCAU (((((.(((((.....))))).....(((((((((.......)))))....((((((((..((...((((((....))))))..))...))))).)))...))))))))).......... ( -41.70) >pfo.3002 5726624 120 - 6438405 CUCACUGGAACCUUGAGUUCCCUGAAGGGCCGUCGAAGACUACGACGUUGAUAGGUUGGGUGUGUAAGCGCUGUGAGGCGUUGAGCUAACCAAUACUAAUUGCCCGUGAGGCUUGACCAU (((((.(((((.....))))).....(((((((((.......)))))....((((((((..((...((((((....))))))..))...))))).)))...))))))))).......... ( -41.70) >pst.2031 3869713 120 - 6397126 CUCACUGGAACCUUGAGUUCCCUGAAGGGCCGUCGAAGACUACGACGUUGAUAGGUUGGGUGUGUAAGCGCUGUGAGGCGUUGAGCUAACCAAUACUAAUUGCCCGUGAGGCUUGACCAU (((((.(((((.....))))).....(((((((((.......)))))....((((((((..((...((((((....))))))..))...))))).)))...))))))))).......... ( -41.70) >consensus CUCACUGGAACCUUGAGUUCCCUGAAGGGCCGUCGAAGACUACGACGUUGAUAGGUUGGGUGUGUAAGCGCUGUGAGGCGUUGAGCUAACCAAUACUAAUUGCCCGUGAGGCUUGACCAU (((((.(((((.....))))).....(((((((((.......)))))....((((((((..((...((((((....))))))..))...))))).)))...))))))))).......... (-43.25 = -41.70 + -1.55)

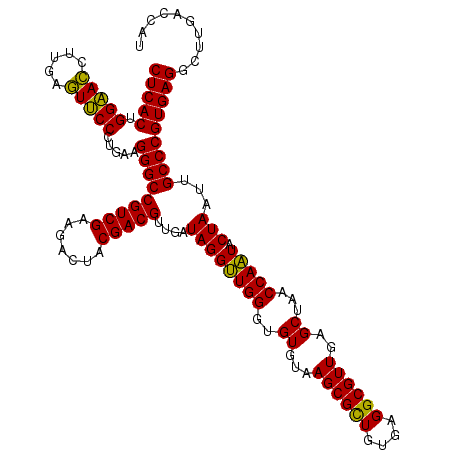
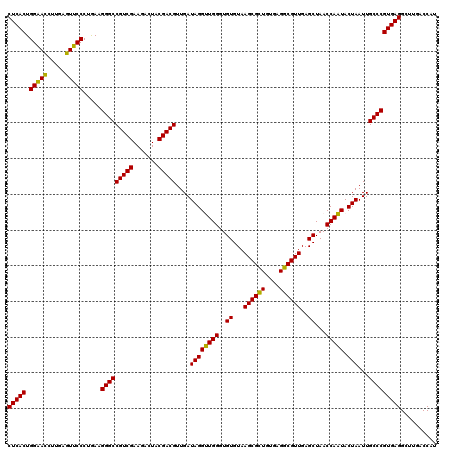
| Location | 6,039,966 – 6,040,086 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.44 |
| Mean single sequence MFE | -40.35 |
| Consensus MFE | -39.93 |
| Energy contribution | -39.93 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953324 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.3201 6039966 120 - 6264404 ACCGGAGUGGACGAACCUCUGGUGUUCCGGUUGUCACGCCAGUGGCAUUGCCGGGUAGCUAUGUUCGGAAAAGAUAACCGCUGAAAGCAUCUAAGCGGGAAACUUGCCUCAAGAUGAGAU (((((((.(......)))))))).(((((((((((..(((...)))....(((..((....))..)))....))))))))..)))..(((((..((((....).)))....))))).... ( -39.90) >pel.69 5768498 120 + 5888780 ACCGGAGUGGACGAACCUCUGGUGUUCCGGUUGUCACGCCAGUGGCAUUGCCGGGUAGCUAUGUUCGGAAGAGAUAACCGCUGAAAGCAUCUAAGCGGGAAACUUGCCUCAAGAUGAGAU (((((((.(......))))))))..((((((((((((....))))))..))))))((((....(((....)))......))))....(((((..((((....).)))....))))).... ( -41.40) >psp.1833 3631837 120 - 5928787 ACCGGAGUGGACGAACCUCUGGUGUUCCGGUUGUCACGCCAGUGGCAUUGCCGGGUAGCUAUGUUCGGAAAAGAUAACCGCUGAAAGCAUCUAAGCGGGAAACUUGCCUCAAGAUGAGAU (((((((.(......)))))))).(((((((((((..(((...)))....(((..((....))..)))....))))))))..)))..(((((..((((....).)))....))))).... ( -39.90) >pf5.3378 6383123 120 - 7074893 ACCGGAGUGGACGAACCUCUGGUGUUCCGGUUGUCACGCCAGUGGCAUUGCCGGGUAGCUAUGUUCGGAAAAGAUAACCGCUGAAAGCAUCUAAGCGGGAAACUUGCCUCAAGAUGAGAU (((((((.(......)))))))).(((((((((((..(((...)))....(((..((....))..)))....))))))))..)))..(((((..((((....).)))....))))).... ( -39.90) >pfo.3002 5726744 120 - 6438405 ACCGGAGUGGACGAACCUCUGGUGUUCCGGUUGUCACGCCAGUGGCAUUGCCGGGUAGCUAUGUUCGGGAAAGAUAACCGCUGAAAGCAUCUAAGCGGGAAACUUGCCUCAAGAUGAGAU (((((((.(......)))))))).(((((((((((..(((...))).((.(((..((....))..))).)).))))))))..)))..(((((..((((....).)))....))))).... ( -41.10) >pst.2031 3869833 120 - 6397126 ACCGGAGUGGACGAACCUCUGGUGUUCCGGUUGUCACGCCAGUGGCAUUGCCGGGUAGCUAUGUUCGGAAAAGAUAACCGCUGAAAGCAUCUAAGCGGGAAACUUGCCUCAAGAUGAGAU (((((((.(......)))))))).(((((((((((..(((...)))....(((..((....))..)))....))))))))..)))..(((((..((((....).)))....))))).... ( -39.90) >consensus ACCGGAGUGGACGAACCUCUGGUGUUCCGGUUGUCACGCCAGUGGCAUUGCCGGGUAGCUAUGUUCGGAAAAGAUAACCGCUGAAAGCAUCUAAGCGGGAAACUUGCCUCAAGAUGAGAU (((((((.(......)))))))).(((((((((((..(((...)))....(((..((....))..)))....))))))))..)))..(((((..((((....).)))....))))).... (-39.93 = -39.93 + -0.00)

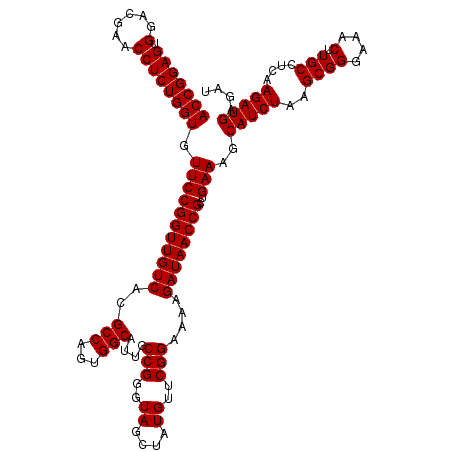
| Location | 6,040,006 – 6,040,126 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.44 |
| Mean single sequence MFE | -42.68 |
| Consensus MFE | -42.71 |
| Energy contribution | -42.57 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.03 |
| Mean z-score | -0.90 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.889679 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.3201 6040006 120 - 6264404 ACGUUUGAGAUUUGAGAGGGGCUGCUCCUAGUACGAGAGGACCGGAGUGGACGAACCUCUGGUGUUCCGGUUGUCACGCCAGUGGCAUUGCCGGGUAGCUAUGUUCGGAAAAGAUAACCG ..........((((((...((((((((.......(.(((.(((((((.(......)))))))).))))(((((((((....))))))..)))))))))))...))))))........... ( -42.70) >pel.69 5768538 120 + 5888780 ACGUUUGAGAUUUGAGAGGGGCUGCUCCUAGUACGAGAGGACCGGAGUGGACGAACCUCUGGUGUUCCGGUUGUCACGCCAGUGGCAUUGCCGGGUAGCUAUGUUCGGAAGAGAUAACCG .................((((((((((.......(.(((.(((((((.(......)))))))).))))(((((((((....))))))..)))))))))))...(((....)))....)). ( -43.20) >psp.1833 3631877 120 - 5928787 ACGUUUGAGAUUUGAGAGGGGCUGCUCCUAGUACGAGAGGACCGGAGUGGACGAACCUCUGGUGUUCCGGUUGUCACGCCAGUGGCAUUGCCGGGUAGCUAUGUUCGGAAAAGAUAACCG ..........((((((...((((((((.......(.(((.(((((((.(......)))))))).))))(((((((((....))))))..)))))))))))...))))))........... ( -42.70) >pf5.3378 6383163 120 - 7074893 ACGUUUGAGAUUUGAGAGGGGCUGCUCCUAGUACGAGAGGACCGGAGUGGACGAACCUCUGGUGUUCCGGUUGUCACGCCAGUGGCAUUGCCGGGUAGCUAUGUUCGGAAAAGAUAACCG ..........((((((...((((((((.......(.(((.(((((((.(......)))))))).))))(((((((((....))))))..)))))))))))...))))))........... ( -42.70) >pfo.3002 5726784 120 - 6438405 ACGUUUGAGAUUUGAGAGGGGCUGCUCCUAGUACGAGAGGACCGGAGUGGACGAACCUCUGGUGUUCCGGUUGUCACGCCAGUGGCAUUGCCGGGUAGCUAUGUUCGGGAAAGAUAACCG ......(((...(.(((((..(..((((..((.(....).)).))))..).....))))).)..)))((((((((..(((...))).((.(((..((....))..))).)).)))))))) ( -42.10) >pst.2031 3869873 120 - 6397126 ACGUUUGAGAUUUGAGAGGGGCUGCUCCUAGUACGAGAGGACCGGAGUGGACGAACCUCUGGUGUUCCGGUUGUCACGCCAGUGGCAUUGCCGGGUAGCUAUGUUCGGAAAAGAUAACCG ..........((((((...((((((((.......(.(((.(((((((.(......)))))))).))))(((((((((....))))))..)))))))))))...))))))........... ( -42.70) >consensus ACGUUUGAGAUUUGAGAGGGGCUGCUCCUAGUACGAGAGGACCGGAGUGGACGAACCUCUGGUGUUCCGGUUGUCACGCCAGUGGCAUUGCCGGGUAGCUAUGUUCGGAAAAGAUAACCG ..........((((((...((((((((.......(.(((.(((((((.(......)))))))).))))(((((((((....))))))..)))))))))))...))))))........... (-42.71 = -42.57 + -0.14)

| Location | 6,040,086 – 6,040,206 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -29.50 |
| Consensus MFE | -29.50 |
| Energy contribution | -29.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.41 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.33 |
| SVM RNA-class probability | 0.992410 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.3201 6040086 120 + 6264404 CCUCUCGUACUAGGAGCAGCCCCUCUCAAAUCUCAAACGUCCACGGCAGAUAGGGACCGAACUGUCUCACGACGUUCUAAACCCAGCUCGCGUACCACUUUAAAUGGCGAACAGCCAUAC ......((((...((((...((((.....((((....((....))..))))))))...((((.(((....)))))))........))))..))))........(((((.....))))).. ( -29.50) >pel.69 5768618 120 - 5888780 CCUCUCGUACUAGGAGCAGCCCCUCUCAAAUCUCAAACGUCCACGGCAGAUAGGGACCGAACUGUCUCACGACGUUCUAAACCCAGCUCGCGUACCACUUUAAAUGGCGAACAGCCAUAC ......((((...((((...((((.....((((....((....))..))))))))...((((.(((....)))))))........))))..))))........(((((.....))))).. ( -29.50) >psp.1833 3631957 120 + 5928787 CCUCUCGUACUAGGAGCAGCCCCUCUCAAAUCUCAAACGUCCACGGCAGAUAGGGACCGAACUGUCUCACGACGUUCUAAACCCAGCUCGCGUACCACUUUAAAUGGCGAACAGCCAUAC ......((((...((((...((((.....((((....((....))..))))))))...((((.(((....)))))))........))))..))))........(((((.....))))).. ( -29.50) >pf5.3378 6383243 120 + 7074893 CCUCUCGUACUAGGAGCAGCCCCUCUCAAAUCUCAAACGUCCACGGCAGAUAGGGACCGAACUGUCUCACGACGUUCUAAACCCAGCUCGCGUACCACUUUAAAUGGCGAACAGCCAUAC ......((((...((((...((((.....((((....((....))..))))))))...((((.(((....)))))))........))))..))))........(((((.....))))).. ( -29.50) >pfo.3002 5726864 120 + 6438405 CCUCUCGUACUAGGAGCAGCCCCUCUCAAAUCUCAAACGUCCACGGCAGAUAGGGACCGAACUGUCUCACGACGUUCUAAACCCAGCUCGCGUACCACUUUAAAUGGCGAACAGCCAUAC ......((((...((((...((((.....((((....((....))..))))))))...((((.(((....)))))))........))))..))))........(((((.....))))).. ( -29.50) >pst.2031 3869953 120 + 6397126 CCUCUCGUACUAGGAGCAGCCCCUCUCAAAUCUCAAACGUCCACGGCAGAUAGGGACCGAACUGUCUCACGACGUUCUAAACCCAGCUCGCGUACCACUUUAAAUGGCGAACAGCCAUAC ......((((...((((...((((.....((((....((....))..))))))))...((((.(((....)))))))........))))..))))........(((((.....))))).. ( -29.50) >consensus CCUCUCGUACUAGGAGCAGCCCCUCUCAAAUCUCAAACGUCCACGGCAGAUAGGGACCGAACUGUCUCACGACGUUCUAAACCCAGCUCGCGUACCACUUUAAAUGGCGAACAGCCAUAC ......((((...((((...((((.....((((....((....))..))))))))...((((.(((....)))))))........))))..))))........(((((.....))))).. (-29.50 = -29.50 + 0.00)

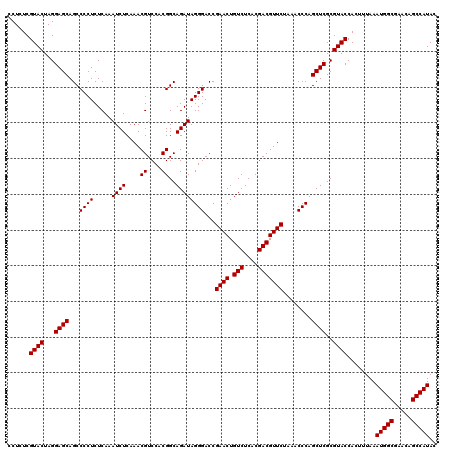
| Location | 6,040,126 – 6,040,246 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -39.50 |
| Consensus MFE | -39.50 |
| Energy contribution | -39.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.28 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.984507 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.3201 6040126 120 + 6264404 CCACGGCAGAUAGGGACCGAACUGUCUCACGACGUUCUAAACCCAGCUCGCGUACCACUUUAAAUGGCGAACAGCCAUACCCUUGGGACCGGCUUCAGCCCCAGGAUGUGAUGAGCCGAC ...((((.....(((...((((.(((....)))))))....)))...((((((.((.......(((((.....))))).((....))...(((....)))...))))))))...)))).. ( -39.50) >pel.69 5768658 120 - 5888780 CCACGGCAGAUAGGGACCGAACUGUCUCACGACGUUCUAAACCCAGCUCGCGUACCACUUUAAAUGGCGAACAGCCAUACCCUUGGGACCGGCUUCAGCCCCAGGAUGUGAUGAGCCGAC ...((((.....(((...((((.(((....)))))))....)))...((((((.((.......(((((.....))))).((....))...(((....)))...))))))))...)))).. ( -39.50) >psp.1833 3631997 120 + 5928787 CCACGGCAGAUAGGGACCGAACUGUCUCACGACGUUCUAAACCCAGCUCGCGUACCACUUUAAAUGGCGAACAGCCAUACCCUUGGGACCGGCUUCAGCCCCAGGAUGUGAUGAGCCGAC ...((((.....(((...((((.(((....)))))))....)))...((((((.((.......(((((.....))))).((....))...(((....)))...))))))))...)))).. ( -39.50) >pf5.3378 6383283 120 + 7074893 CCACGGCAGAUAGGGACCGAACUGUCUCACGACGUUCUAAACCCAGCUCGCGUACCACUUUAAAUGGCGAACAGCCAUACCCUUGGGACCGGCUUCAGCCCCAGGAUGUGAUGAGCCGAC ...((((.....(((...((((.(((....)))))))....)))...((((((.((.......(((((.....))))).((....))...(((....)))...))))))))...)))).. ( -39.50) >pfo.3002 5726904 120 + 6438405 CCACGGCAGAUAGGGACCGAACUGUCUCACGACGUUCUAAACCCAGCUCGCGUACCACUUUAAAUGGCGAACAGCCAUACCCUUGGGACCGGCUUCAGCCCCAGGAUGUGAUGAGCCGAC ...((((.....(((...((((.(((....)))))))....)))...((((((.((.......(((((.....))))).((....))...(((....)))...))))))))...)))).. ( -39.50) >pst.2031 3869993 120 + 6397126 CCACGGCAGAUAGGGACCGAACUGUCUCACGACGUUCUAAACCCAGCUCGCGUACCACUUUAAAUGGCGAACAGCCAUACCCUUGGGACCGGCUUCAGCCCCAGGAUGUGAUGAGCCGAC ...((((.....(((...((((.(((....)))))))....)))...((((((.((.......(((((.....))))).((....))...(((....)))...))))))))...)))).. ( -39.50) >consensus CCACGGCAGAUAGGGACCGAACUGUCUCACGACGUUCUAAACCCAGCUCGCGUACCACUUUAAAUGGCGAACAGCCAUACCCUUGGGACCGGCUUCAGCCCCAGGAUGUGAUGAGCCGAC ...((((.....(((...((((.(((....)))))))....)))...((((((.((.......(((((.....))))).((....))...(((....)))...))))))))...)))).. (-39.50 = -39.50 + 0.00)

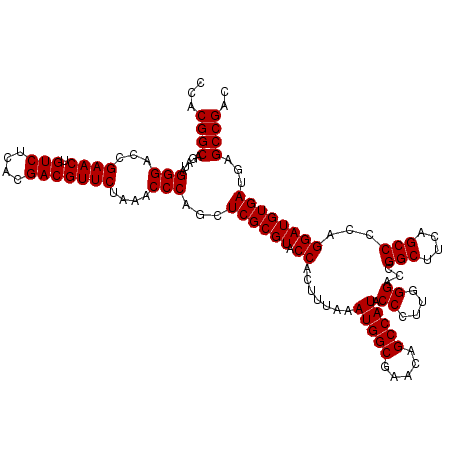
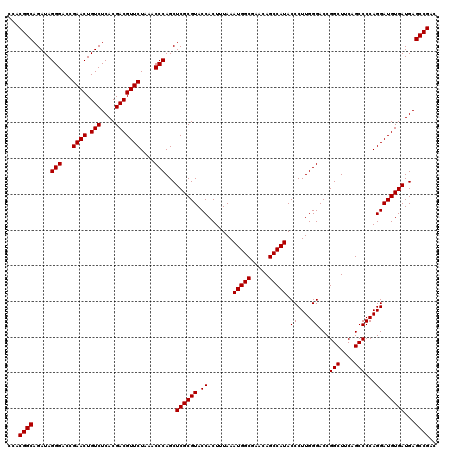
| Location | 6,040,126 – 6,040,246 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -50.70 |
| Consensus MFE | -50.70 |
| Energy contribution | -50.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.10 |
| Structure conservation index | 1.00 |
| SVM decision value | 3.92 |
| SVM RNA-class probability | 0.999708 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.3201 6040126 120 - 6264404 GUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUUAAAGUGGUACGCGAGCUGGGUUUAGAACGUCGUGAGACAGUUCGGUCCCUAUCUGCCGUGG ..((((..(((((((((((((((((....))))))).)))))(((((.....))))).....)))))....((...(((....(((((((....))).))))...)))..)).))))... ( -50.70) >pel.69 5768658 120 + 5888780 GUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUUAAAGUGGUACGCGAGCUGGGUUUAGAACGUCGUGAGACAGUUCGGUCCCUAUCUGCCGUGG ..((((..(((((((((((((((((....))))))).)))))(((((.....))))).....)))))....((...(((....(((((((....))).))))...)))..)).))))... ( -50.70) >psp.1833 3631997 120 - 5928787 GUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUUAAAGUGGUACGCGAGCUGGGUUUAGAACGUCGUGAGACAGUUCGGUCCCUAUCUGCCGUGG ..((((..(((((((((((((((((....))))))).)))))(((((.....))))).....)))))....((...(((....(((((((....))).))))...)))..)).))))... ( -50.70) >pf5.3378 6383283 120 - 7074893 GUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUUAAAGUGGUACGCGAGCUGGGUUUAGAACGUCGUGAGACAGUUCGGUCCCUAUCUGCCGUGG ..((((..(((((((((((((((((....))))))).)))))(((((.....))))).....)))))....((...(((....(((((((....))).))))...)))..)).))))... ( -50.70) >pfo.3002 5726904 120 - 6438405 GUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUUAAAGUGGUACGCGAGCUGGGUUUAGAACGUCGUGAGACAGUUCGGUCCCUAUCUGCCGUGG ..((((..(((((((((((((((((....))))))).)))))(((((.....))))).....)))))....((...(((....(((((((....))).))))...)))..)).))))... ( -50.70) >pst.2031 3869993 120 - 6397126 GUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUUAAAGUGGUACGCGAGCUGGGUUUAGAACGUCGUGAGACAGUUCGGUCCCUAUCUGCCGUGG ..((((..(((((((((((((((((....))))))).)))))(((((.....))))).....)))))....((...(((....(((((((....))).))))...)))..)).))))... ( -50.70) >consensus GUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUUAAAGUGGUACGCGAGCUGGGUUUAGAACGUCGUGAGACAGUUCGGUCCCUAUCUGCCGUGG ..((((..(((((((((((((((((....))))))).)))))(((((.....))))).....)))))....((...(((....(((((((....))).))))...)))..)).))))... (-50.70 = -50.70 + -0.00)

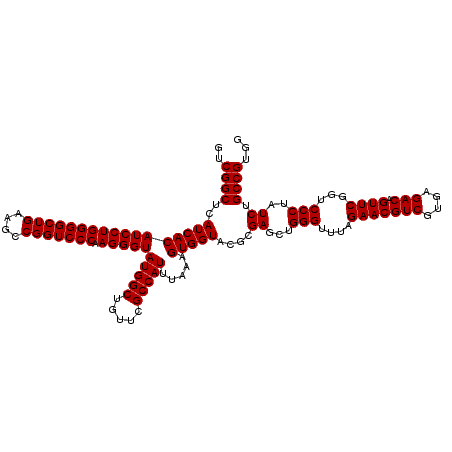
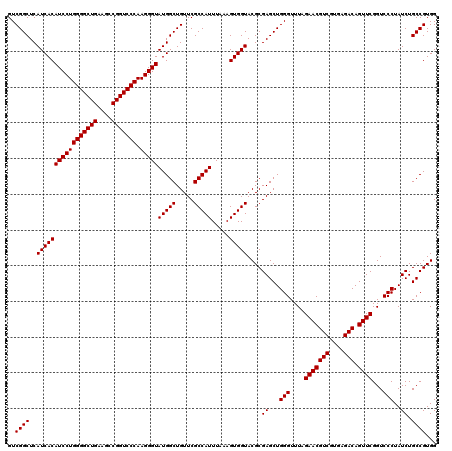
| Location | 6,040,166 – 6,040,286 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -51.40 |
| Consensus MFE | -51.40 |
| Energy contribution | -51.40 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.36 |
| Structure conservation index | 1.00 |
| SVM decision value | 4.38 |
| SVM RNA-class probability | 0.999884 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.3201 6040166 120 - 6264404 CCCAAGAGUUCAUAUCGACGGCGGUGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUUAAAGUGGUACGCGAGCUGGGU ((((.((((..(((((((....((((.....)))))))))))..))))(((((((((((((((((....))))))).)))))(((((.....))))).....)))))........)))). ( -51.40) >pel.69 5768698 120 + 5888780 CCCAAGAGUUCAUAUCGACGGCGGUGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUUAAAGUGGUACGCGAGCUGGGU ((((.((((..(((((((....((((.....)))))))))))..))))(((((((((((((((((....))))))).)))))(((((.....))))).....)))))........)))). ( -51.40) >psp.1833 3632037 120 - 5928787 CCCAAGAGUUCAUAUCGACGGCGGUGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUUAAAGUGGUACGCGAGCUGGGU ((((.((((..(((((((....((((.....)))))))))))..))))(((((((((((((((((....))))))).)))))(((((.....))))).....)))))........)))). ( -51.40) >pf5.3378 6383323 120 - 7074893 CCCAAGAGUUCAUAUCGACGGCGGUGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUUAAAGUGGUACGCGAGCUGGGU ((((.((((..(((((((....((((.....)))))))))))..))))(((((((((((((((((....))))))).)))))(((((.....))))).....)))))........)))). ( -51.40) >pfo.3002 5726944 120 - 6438405 CCCAAGAGUUCAUAUCGACGGCGGUGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUUAAAGUGGUACGCGAGCUGGGU ((((.((((..(((((((....((((.....)))))))))))..))))(((((((((((((((((....))))))).)))))(((((.....))))).....)))))........)))). ( -51.40) >pst.2031 3870033 120 - 6397126 CCCAAGAGUUCAUAUCGACGGCGGUGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUUAAAGUGGUACGCGAGCUGGGU ((((.((((..(((((((....((((.....)))))))))))..))))(((((((((((((((((....))))))).)))))(((((.....))))).....)))))........)))). ( -51.40) >consensus CCCAAGAGUUCAUAUCGACGGCGGUGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUUAAAGUGGUACGCGAGCUGGGU ((((.((((..(((((((....((((.....)))))))))))..))))(((((((((((((((((....))))))).)))))(((((.....))))).....)))))........)))). (-51.40 = -51.40 + -0.00)
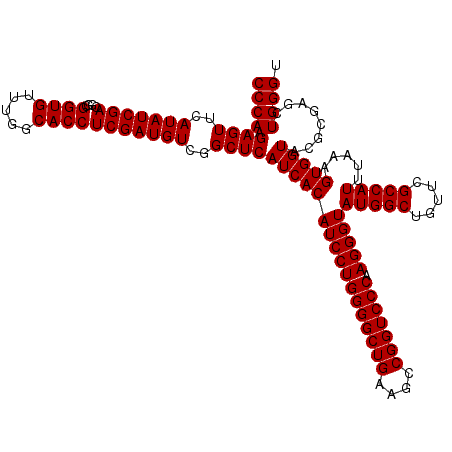
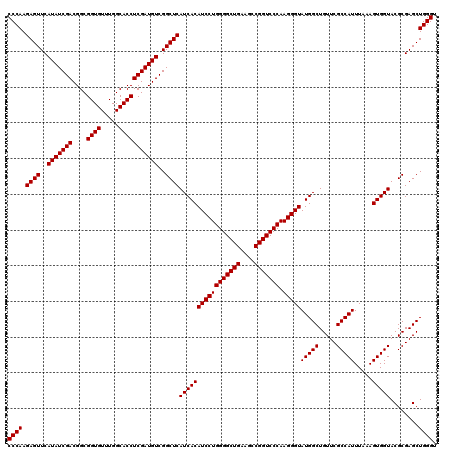
| Location | 6,040,206 – 6,040,326 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -44.20 |
| Consensus MFE | -44.20 |
| Energy contribution | -44.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.35 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.16 |
| SVM RNA-class probability | 0.989397 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.3201 6040206 120 - 6264404 AACGGAUAAAAGGUACUCCGGGGAUAACAGGCUGAUACCGCCCAAGAGUUCAUAUCGACGGCGGUGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGG ..((((..........))))(((((....((((.((((((((...((.......))...))))))))(..((.((..(((((.((....)))))))..)).))..))))).))))).... ( -44.20) >pel.69 5768738 120 + 5888780 AACGGAUAAAAGGUACUCCGGGGAUAACAGGCUGAUACCGCCCAAGAGUUCAUAUCGACGGCGGUGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGG ..((((..........))))(((((....((((.((((((((...((.......))...))))))))(..((.((..(((((.((....)))))))..)).))..))))).))))).... ( -44.20) >psp.1833 3632077 120 - 5928787 AACGGAUAAAAGGUACUCCGGGGAUAACAGGCUGAUACCGCCCAAGAGUUCAUAUCGACGGCGGUGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGG ..((((..........))))(((((....((((.((((((((...((.......))...))))))))(..((.((..(((((.((....)))))))..)).))..))))).))))).... ( -44.20) >pf5.3378 6383363 120 - 7074893 AACGGAUAAAAGGUACUCCGGGGAUAACAGGCUGAUACCGCCCAAGAGUUCAUAUCGACGGCGGUGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGG ..((((..........))))(((((....((((.((((((((...((.......))...))))))))(..((.((..(((((.((....)))))))..)).))..))))).))))).... ( -44.20) >pfo.3002 5726984 120 - 6438405 AACGGAUAAAAGGUACUCCGGGGAUAACAGGCUGAUACCGCCCAAGAGUUCAUAUCGACGGCGGUGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGG ..((((..........))))(((((....((((.((((((((...((.......))...))))))))(..((.((..(((((.((....)))))))..)).))..))))).))))).... ( -44.20) >pst.2031 3870073 120 - 6397126 AACGGAUAAAAGGUACUCCGGGGAUAACAGGCUGAUACCGCCCAAGAGUUCAUAUCGACGGCGGUGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGG ..((((..........))))(((((....((((.((((((((...((.......))...))))))))(..((.((..(((((.((....)))))))..)).))..))))).))))).... ( -44.20) >consensus AACGGAUAAAAGGUACUCCGGGGAUAACAGGCUGAUACCGCCCAAGAGUUCAUAUCGACGGCGGUGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGG ..((((..........))))(((((....((((.((((((((...((.......))...))))))))(..((.((..(((((.((....)))))))..)).))..))))).))))).... (-44.20 = -44.20 + -0.00)

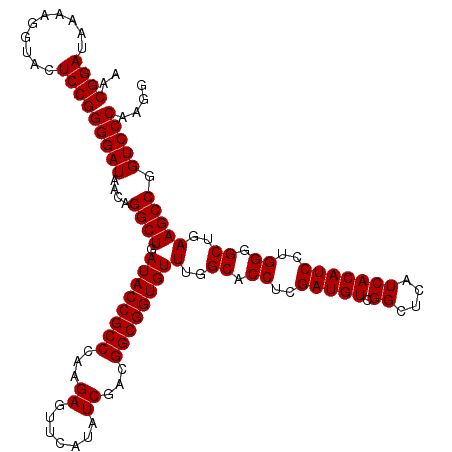
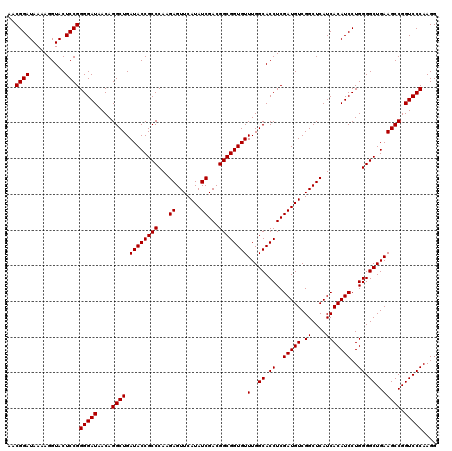
| Location | 6,040,246 – 6,040,366 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -40.90 |
| Consensus MFE | -40.90 |
| Energy contribution | -40.90 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.90 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.874804 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.3201 6040246 120 - 6264404 CUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGGUACUCCGGGGAUAACAGGCUGAUACCGCCCAAGAGUUCAUAUCGACGGCGGUGUUUGGCACCUCGAU ....((.(((((((((((((.......)))))))))......((((..........)))).)))).))..((..((((((((...((.......))...))))))).)..))........ ( -40.90) >pel.69 5768778 120 + 5888780 CUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGGUACUCCGGGGAUAACAGGCUGAUACCGCCCAAGAGUUCAUAUCGACGGCGGUGUUUGGCACCUCGAU ....((.(((((((((((((.......)))))))))......((((..........)))).)))).))..((..((((((((...((.......))...))))))).)..))........ ( -40.90) >psp.1833 3632117 120 - 5928787 CUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGGUACUCCGGGGAUAACAGGCUGAUACCGCCCAAGAGUUCAUAUCGACGGCGGUGUUUGGCACCUCGAU ....((.(((((((((((((.......)))))))))......((((..........)))).)))).))..((..((((((((...((.......))...))))))).)..))........ ( -40.90) >pf5.3378 6383403 120 - 7074893 CUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGGUACUCCGGGGAUAACAGGCUGAUACCGCCCAAGAGUUCAUAUCGACGGCGGUGUUUGGCACCUCGAU ....((.(((((((((((((.......)))))))))......((((..........)))).)))).))..((..((((((((...((.......))...))))))).)..))........ ( -40.90) >pfo.3002 5727024 120 - 6438405 CUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGGUACUCCGGGGAUAACAGGCUGAUACCGCCCAAGAGUUCAUAUCGACGGCGGUGUUUGGCACCUCGAU ....((.(((((((((((((.......)))))))))......((((..........)))).)))).))..((..((((((((...((.......))...))))))).)..))........ ( -40.90) >pst.2031 3870113 120 - 6397126 CUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGGUACUCCGGGGAUAACAGGCUGAUACCGCCCAAGAGUUCAUAUCGACGGCGGUGUUUGGCACCUCGAU ....((.(((((((((((((.......)))))))))......((((..........)))).)))).))..((..((((((((...((.......))...))))))).)..))........ ( -40.90) >consensus CUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGGUACUCCGGGGAUAACAGGCUGAUACCGCCCAAGAGUUCAUAUCGACGGCGGUGUUUGGCACCUCGAU ....((.(((((((((((((.......)))))))))......((((..........)))).)))).))..((..((((((((...((.......))...))))))).)..))........ (-40.90 = -40.90 + -0.00)

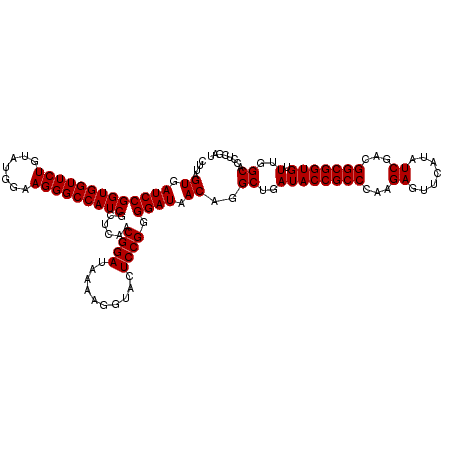
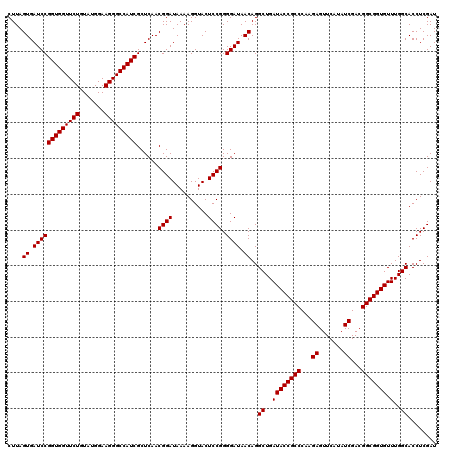
| Location | 6,040,286 – 6,040,406 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.72 |
| Mean single sequence MFE | -42.70 |
| Consensus MFE | -42.70 |
| Energy contribution | -42.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.15 |
| Structure conservation index | 1.00 |
| SVM decision value | 4.10 |
| SVM RNA-class probability | 0.999794 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.3201 6040286 120 - 6264404 GACUGCGAGACAGACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGGUACUCCGGGGAUAACAGGCUGAUACCG ..((((..(((......)))..)))).(((....)))(((.(((((.(((((((((((((.......)))))))))......((((..........)))).)))).....))))).))). ( -42.70) >pel.69 5768818 120 + 5888780 GACUGCGAGACAAACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGGUACUCCGGGGAUAACAGGCUGAUACCG ..((((..(((......)))..)))).(((....)))(((.(((((.(((((((((((((.......)))))))))......((((..........)))).)))).....))))).))). ( -42.70) >psp.1833 3632157 120 - 5928787 GACUGCGAGACAGACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGGUACUCCGGGGAUAACAGGCUGAUACCG ..((((..(((......)))..)))).(((....)))(((.(((((.(((((((((((((.......)))))))))......((((..........)))).)))).....))))).))). ( -42.70) >pf5.3378 6383443 120 - 7074893 GACUGCGAGACAGACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGGUACUCCGGGGAUAACAGGCUGAUACCG ..((((..(((......)))..)))).(((....)))(((.(((((.(((((((((((((.......)))))))))......((((..........)))).)))).....))))).))). ( -42.70) >pfo.3002 5727064 120 - 6438405 GACUGCGAGACAGACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGGUACUCCGGGGAUAACAGGCUGAUACCG ..((((..(((......)))..)))).(((....)))(((.(((((.(((((((((((((.......)))))))))......((((..........)))).)))).....))))).))). ( -42.70) >pst.2031 3870153 120 - 6397126 GACUGCGAGACAGACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGGUACUCCGGGGAUAACAGGCUGAUACCG ..((((..(((......)))..)))).(((....)))(((.(((((.(((((((((((((.......)))))))))......((((..........)))).)))).....))))).))). ( -42.70) >consensus GACUGCGAGACAGACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGGUACUCCGGGGAUAACAGGCUGAUACCG ..((((..(((......)))..)))).(((....)))(((.(((((.(((((((((((((.......)))))))))......((((..........)))).)))).....))))).))). (-42.70 = -42.70 + -0.00)

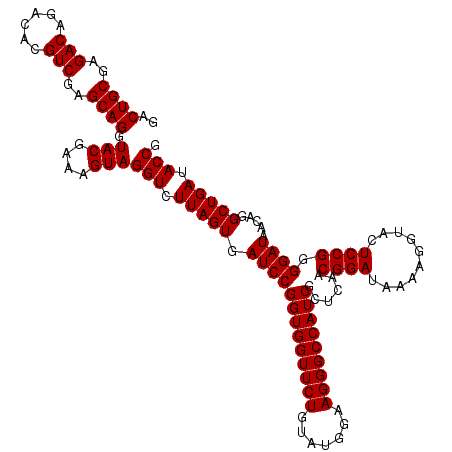
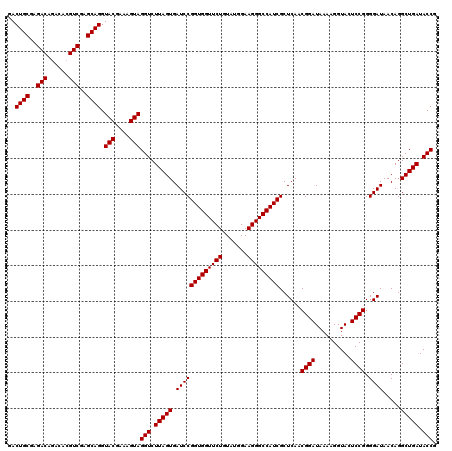
| Location | 6,040,366 – 6,040,486 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.00 |
| Mean single sequence MFE | -28.10 |
| Consensus MFE | -27.83 |
| Energy contribution | -27.83 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.65 |
| SVM RNA-class probability | 0.996052 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.3201 6040366 120 + 6264404 ACCUACUUUCGUACCUGCUCGACGUGUCUGUCUCGCAGUCAAGCGCGCUUUUGCCUUUAUACUCUGCGACCGAUUUCCGACCGGUCUGAGCGCACCUUCGUACUCCUCCGUUACUCUUUA ..........((((..(((.((((((.......))).))).)))((((((..((...........))(((((.........))))).))))))......))))................. ( -28.50) >pau.3340 6313285 120 + 6537648 ACCUACUUUCGUACCUGCUCGACGUGUCUGUCUCGCAGUCAAGCGCGCUUUUGCCUUUAUACUCUGCGACCGAUUUCCGACCGGUCUGAGCGCACCUUCGUACUCCUCCGUUACUCUUUA ..........((((..(((.((((((.......))).))).)))((((((..((...........))(((((.........))))).))))))......))))................. ( -28.50) >pel.69 5768898 120 - 5888780 ACCUACUUUCGUACCUGCUCGACGUGUUUGUCUCGCAGUCAAGCGCGCUUUUGCCUUUAUACUCUACGACCGAUUUCCGACCGGUCUGAGCGCACCUUCGUACUCCUCCGUUACUCUUUG ..........((((((((..((((....))))..))))....((((((....)).............(((((.........)))))...))))......))))................. ( -28.50) >pf5.3378 6383523 120 + 7074893 ACCUACUUUCGUACCUGCUCGACGUGUCUGUCUCGCAGUCAAGCGCGCUUUUGCCUUUAUACUCUACGACCGAUUUCCGACCGGUCUGAGCGCACCUUCGUACUCCUCCGUUACUCUUUA ..........((((((((..((((....))))..))))....((((((....)).............(((((.........)))))...))))......))))................. ( -27.70) >pfo.3002 5727144 120 + 6438405 ACCUACUUUCGUACCUGCUCGACGUGUCUGUCUCGCAGUCAAGCGCGCUUUUGCCUUUAUACUCUACGACCGAUUUCCGACCGGUCUGAGCGCACCUUCGUACUCCUCCGUUACUCUUUA ..........((((((((..((((....))))..))))....((((((....)).............(((((.........)))))...))))......))))................. ( -27.70) >pst.2031 3870233 120 + 6397126 ACCUACUUUCGUACCUGCUCGACGUGUCUGUCUCGCAGUCAAGCGCGCUUUUGCCUUUAUACUCUACGACCGAUUUCCGACCGGUCUGAGCGCACCUUCGUACUCCUCCGUUACUCUUUA ..........((((((((..((((....))))..))))....((((((....)).............(((((.........)))))...))))......))))................. ( -27.70) >consensus ACCUACUUUCGUACCUGCUCGACGUGUCUGUCUCGCAGUCAAGCGCGCUUUUGCCUUUAUACUCUACGACCGAUUUCCGACCGGUCUGAGCGCACCUUCGUACUCCUCCGUUACUCUUUA ..........((((((((..((((....))))..))))....((((((....)).............(((((.........)))))...))))......))))................. (-27.83 = -27.83 + -0.00)

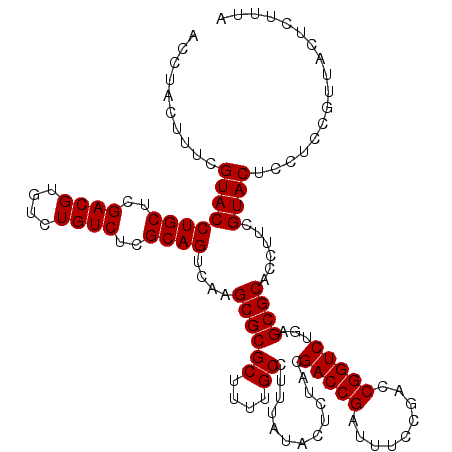
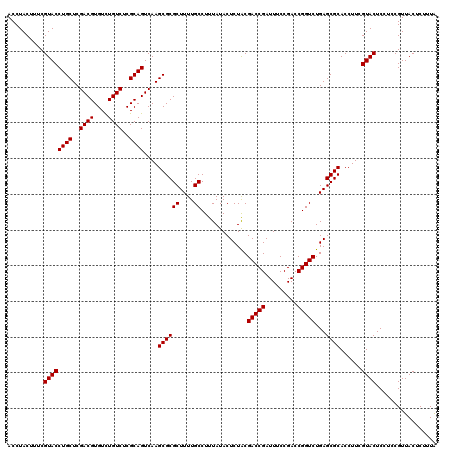
| Location | 6,040,366 – 6,040,486 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.00 |
| Mean single sequence MFE | -38.70 |
| Consensus MFE | -38.50 |
| Energy contribution | -38.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.99 |
| SVM decision value | 4.90 |
| SVM RNA-class probability | 0.999960 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.3201 6040366 120 - 6264404 UAAAGAGUAACGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGCAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGAGCAGGUACGAAAGUAGGU .................((((..(((((((((..(((((((.....)))))))((...........))...)))))))))..((((..(((......)))..)))))))).......... ( -39.10) >pau.3340 6313285 120 - 6537648 UAAAGAGUAACGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGCAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGAGCAGGUACGAAAGUAGGU .................((((..(((((((((..(((((((.....)))))))((...........))...)))))))))..((((..(((......)))..)))))))).......... ( -39.10) >pel.69 5768898 120 + 5888780 CAAAGAGUAACGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAAACACGUCGAGCAGGUACGAAAGUAGGU .................((((..(((((((((..(((((((.....))))))).....((......))...)))))))))..((((..(((......)))..)))))))).......... ( -38.50) >pf5.3378 6383523 120 - 7074893 UAAAGAGUAACGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGAGCAGGUACGAAAGUAGGU .................((((..(((((((((..(((((((.....))))))).....((......))...)))))))))..((((..(((......)))..)))))))).......... ( -38.50) >pfo.3002 5727144 120 - 6438405 UAAAGAGUAACGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGAGCAGGUACGAAAGUAGGU .................((((..(((((((((..(((((((.....))))))).....((......))...)))))))))..((((..(((......)))..)))))))).......... ( -38.50) >pst.2031 3870233 120 - 6397126 UAAAGAGUAACGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGAGCAGGUACGAAAGUAGGU .................((((..(((((((((..(((((((.....))))))).....((......))...)))))))))..((((..(((......)))..)))))))).......... ( -38.50) >consensus UAAAGAGUAACGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGAGCAGGUACGAAAGUAGGU .................((((..(((((((((..(((((((.....))))))).....((......))...)))))))))..((((..(((......)))..)))))))).......... (-38.50 = -38.50 + 0.00)

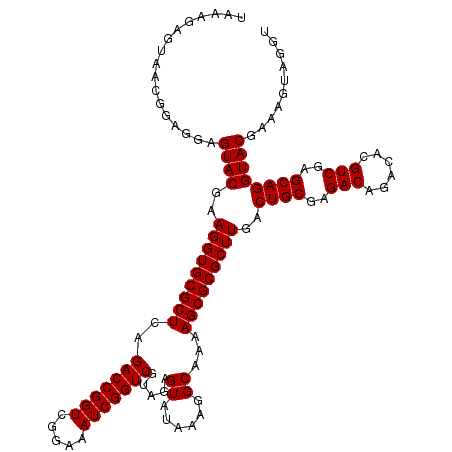
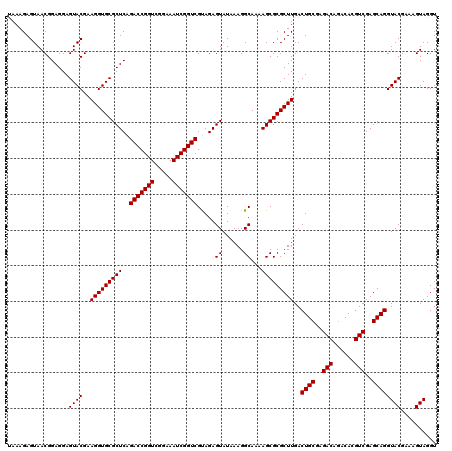
| Location | 6,040,406 – 6,040,526 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.83 |
| Mean single sequence MFE | -30.49 |
| Consensus MFE | -29.73 |
| Energy contribution | -29.73 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.48 |
| SVM RNA-class probability | 0.994428 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.3201 6040406 120 + 6264404 AAGCGCGCUUUUGCCUUUAUACUCUGCGACCGAUUUCCGACCGGUCUGAGCGCACCUUCGUACUCCUCCGUUACUCUUUAGGAGGAGACCGCCCCAGUCAAACUGCCCACCAUACACUGU ..((((((((..((...........))(((((.........))))).))))))......((.(((((((...........))))))))).))..(((.....)))............... ( -32.10) >pau.3340 6313325 120 + 6537648 AAGCGCGCUUUUGCCUUUAUACUCUGCGACCGAUUUCCGACCGGUCUGAGCGCACCUUCGUACUCCUCCGUUACUCUUUAGGAGGAGACCGCCCCAGUCAAACUGCCCACCAUACACUGU ..((((((((..((...........))(((((.........))))).))))))......((.(((((((...........))))))))).))..(((.....)))............... ( -32.10) >pel.69 5768938 120 - 5888780 AAGCGCGCUUUUGCCUUUAUACUCUACGACCGAUUUCCGACCGGUCUGAGCGCACCUUCGUACUCCUCCGUUACUCUUUGGGAGGAGACCGCCCCAGUCAAACUACCCACCAUACACUGU ..((((((....)).............(((((.........)))))...)))).....((..(((((((...........)))))))..))...((((.................)))). ( -29.23) >pf5.3378 6383563 120 + 7074893 AAGCGCGCUUUUGCCUUUAUACUCUACGACCGAUUUCCGACCGGUCUGAGCGCACCUUCGUACUCCUCCGUUACUCUUUAGGAGGAGACCGCCCCAGUCAAACUACCCACCAUACACUGU ..((((((....)).............(((((.........)))))...)))).....((..(((((((...........)))))))..))...((((.................)))). ( -29.83) >pfo.3002 5727184 120 + 6438405 AAGCGCGCUUUUGCCUUUAUACUCUACGACCGAUUUCCGACCGGUCUGAGCGCACCUUCGUACUCCUCCGUUACUCUUUAGGAGGAGACCGCCCCAGUCAAACUACCCACCAUACACUGU ..((((((....)).............(((((.........)))))...)))).....((..(((((((...........)))))))..))...((((.................)))). ( -29.83) >pst.2031 3870273 120 + 6397126 AAGCGCGCUUUUGCCUUUAUACUCUACGACCGAUUUCCGACCGGUCUGAGCGCACCUUCGUACUCCUCCGUUACUCUUUAGGAGGAGACCGCCCCAGUCAAACUACCCACCAUACACUGU ..((((((....)).............(((((.........)))))...)))).....((..(((((((...........)))))))..))...((((.................)))). ( -29.83) >consensus AAGCGCGCUUUUGCCUUUAUACUCUACGACCGAUUUCCGACCGGUCUGAGCGCACCUUCGUACUCCUCCGUUACUCUUUAGGAGGAGACCGCCCCAGUCAAACUACCCACCAUACACUGU ..((((((....)).............(((((.........)))))...)))).....((..(((((((...........)))))))..))...((((.................)))). (-29.73 = -29.73 + 0.00)

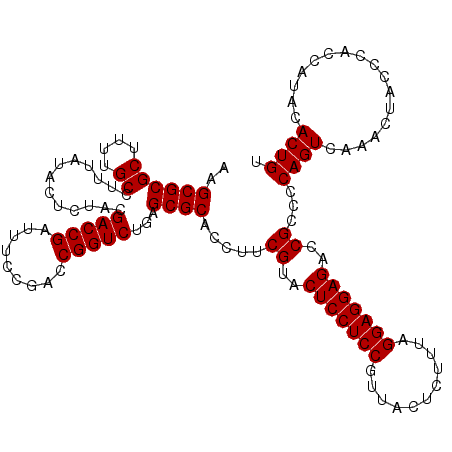
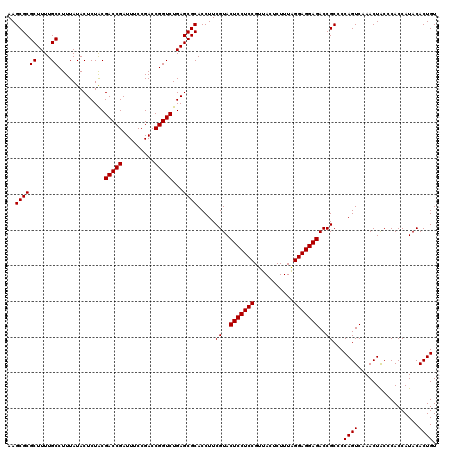
| Location | 6,040,406 – 6,040,526 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.83 |
| Mean single sequence MFE | -39.72 |
| Consensus MFE | -39.77 |
| Energy contribution | -39.55 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.22 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.985662 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.3201 6040406 120 - 6264404 ACAGUGUAUGGUGGGCAGUUUGACUGGGGCGGUCUCCUCCUAAAGAGUAACGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGCAGAGUAUAAAGGCAAAAGCGCGCUU .........((((.((.......((((((((..(((((((...........)))))))(((....)))))))).(((((((.....))))))).))).((......))....)).)))). ( -41.20) >pau.3340 6313325 120 - 6537648 ACAGUGUAUGGUGGGCAGUUUGACUGGGGCGGUCUCCUCCUAAAGAGUAACGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGCAGAGUAUAAAGGCAAAAGCGCGCUU .........((((.((.......((((((((..(((((((...........)))))))(((....)))))))).(((((((.....))))))).))).((......))....)).)))). ( -41.20) >pel.69 5768938 120 + 5888780 ACAGUGUAUGGUGGGUAGUUUGACUGGGGCGGUCUCCUCCCAAAGAGUAACGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAUAAAGGCAAAAGCGCGCUU .........((((.((..((((.((.(((((..(((((((...........)))))))(((....)))))))).(((((((.....)))))))............)))))).)).)))). ( -38.60) >pf5.3378 6383563 120 - 7074893 ACAGUGUAUGGUGGGUAGUUUGACUGGGGCGGUCUCCUCCUAAAGAGUAACGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAUAAAGGCAAAAGCGCGCUU .........((((.((..((((.((.(((((..(((((((...........)))))))(((....)))))))).(((((((.....)))))))............)))))).)).)))). ( -39.10) >pfo.3002 5727184 120 - 6438405 ACAGUGUAUGGUGGGUAGUUUGACUGGGGCGGUCUCCUCCUAAAGAGUAACGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAUAAAGGCAAAAGCGCGCUU .........((((.((..((((.((.(((((..(((((((...........)))))))(((....)))))))).(((((((.....)))))))............)))))).)).)))). ( -39.10) >pst.2031 3870273 120 - 6397126 ACAGUGUAUGGUGGGUAGUUUGACUGGGGCGGUCUCCUCCUAAAGAGUAACGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAUAAAGGCAAAAGCGCGCUU .........((((.((..((((.((.(((((..(((((((...........)))))))(((....)))))))).(((((((.....)))))))............)))))).)).)))). ( -39.10) >consensus ACAGUGUAUGGUGGGUAGUUUGACUGGGGCGGUCUCCUCCUAAAGAGUAACGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAUAAAGGCAAAAGCGCGCUU .........((((.((..((((.((.(((((..(((((((...........)))))))(((....)))))))).(((((((.....)))))))............)))))).)).)))). (-39.77 = -39.55 + -0.22)

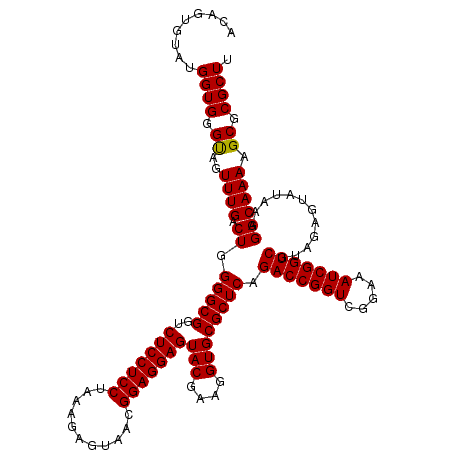
Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:03:45 2007