
| Sequence ID | pao.2545 |
|---|---|
| Location | 4,789,810 – 4,790,848 |
| Length | 1038 |
| Max. P | 0.999978 |

| Location | 4,789,810 – 4,789,930 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.67 |
| Mean single sequence MFE | -46.70 |
| Consensus MFE | -46.70 |
| Energy contribution | -46.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.16 |
| Structure conservation index | 1.00 |
| SVM decision value | 4.35 |
| SVM RNA-class probability | 0.999878 |
| Prediction | RNA |
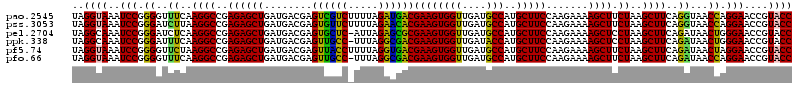
Download alignment: ClustalW | MAF
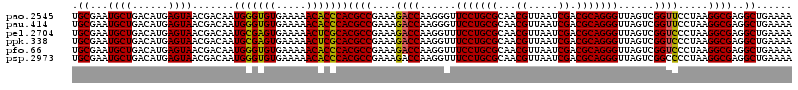
>pao.2545 4789810 120 - 6264404 AAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCGCAAGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGGCGGCCGUAACUAUAACGGUCCUAA ....((((.(((((......(((.(((((...((((((..((((..(((((((((((.((....))..))))))..)))))))))))))))...))))).)))..)))))...))))... ( -47.30) >pel.2704 4999728 120 - 5888780 AAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCGCAAGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGGCGGCCGUAACUAUAACGGUCCUAA ....((((.(((((......(((.(((((...((((((..((((..(((((((((((.((....))..))))))..)))))))))))))))...))))).)))..)))))...))))... ( -47.30) >ppk.338 5480098 120 + 6181863 AAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCGCAAGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGGCGGCCGUAACUAUAACGGUCCUAA ....((((.(((((......(((.(((((...((((((..((((..(((((((((((.((....))..))))))..)))))))))))))))...))))).)))..)))))...))))... ( -47.30) >pf5.74 6948109 120 + 7074893 AAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCUAACGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGGCGGCCGUAACUAUAACGGUCCUAA ....((((.(((((......(((.(((((...((((((..((((..(((((((((((.((....))..))))))..)))))))))))))))...))))).)))..)))))...))))... ( -45.50) >pfo.66 6312979 120 + 6438405 AAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCUAACGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGGCGGCCGUAACUAUAACGGUCCUAA ....((((.(((((......(((.(((((...((((((..((((..(((((((((((.((....))..))))))..)))))))))))))))...))))).)))..)))))...))))... ( -45.50) >psp.2973 5780363 120 - 5928787 AAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCGCAAGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGGCGGCCGUAACUAUAACGGUCCUAA ....((((.(((((......(((.(((((...((((((..((((..(((((((((((.((....))..))))))..)))))))))))))))...))))).)))..)))))...))))... ( -47.30) >consensus AAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCGCAAGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGGCGGCCGUAACUAUAACGGUCCUAA ....((((.(((((......(((.(((((...((((((..((((..(((((((((((.((....))..))))))..)))))))))))))))...))))).)))..)))))...))))... (-46.70 = -46.70 + -0.00)


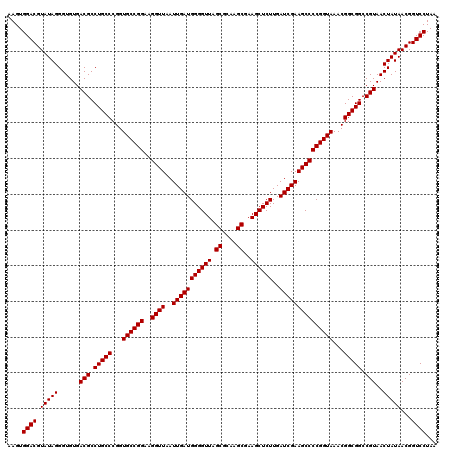
| Location | 4,789,930 – 4,790,050 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.56 |
| Mean single sequence MFE | -39.05 |
| Consensus MFE | -38.58 |
| Energy contribution | -37.05 |
| Covariance contribution | -1.53 |
| Combinations/Pair | 1.17 |
| Mean z-score | -0.86 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.828559 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4789930 120 - 6264404 CGUAACUUCGGGAGAAGGUGCGCCGGCUAGGGUGAAGGAUUUACUCCGUAAGCUCUGGCUGGUCGAAGAUACCAGGCCGCUGCGACUGUUUAUUAAAAACACAGCACUCUGCAAACACGA .....((((....))))((((((((((((((((...(((.....)))....))))))))))).)).......((((..((((....(((((.....)))))))))..))))....))).. ( -40.40) >pau.414 5800761 120 + 6537648 CGUAACUUCGGGAGAAGGUGCGCCGGCUAGGGUGAAGGAUUUACUCCGUAAGCUCUGGCUGGUCGAAGAUACCAGGCCGCUGCGACUGUUUAUUAAAAACACAGCACUCUGCAAACACGA .....((((....))))((((((((((((((((...(((.....)))....))))))))))).)).......((((..((((....(((((.....)))))))))..))))....))).. ( -40.40) >pss.3053 5949317 120 - 6093698 CGUAACUUCGGGAGAAGGUGCGCCGGUGGAGGUGAAGCAUUUACUGCGUAAGCCCCUGCCGGUCGAAGAUACCAGGCCGCUGCGACUGUUUAUUAAAAACACAGCACUCUGCAAACACGA .....((((....))))(((((((((..(.(((...(((.....)))....))).)..)))).)).......((((..((((....(((((.....)))))))))..))))....))).. ( -37.60) >ppk.338 5480218 120 + 6181863 CGUAACUUCGGGAGAAGGUGCGCCGGCGAGGGUGAAGGACUUGCUCCGUAAGCCCAUGCCGGUCGAAGAUACCAGGCCGCUGCGACUGUUUAUUAAAAACACAGCACUCUGCAAACACGA .....((((....))))(((((((((((.((((...(((.....)))....)))).)))))).)).......((((..((((....(((((.....)))))))))..))))....))).. ( -40.60) >pf5.74 6948229 120 + 7074893 CGUAACUUCGGGAGAAGGUGCGCCGGUGAGGGUGAAGGACUUGCUCCGUAAGCUCAUGCCGGUCGAAGAUACCAGGCCGCUGCGACUGUUUAUUAAAAACACAGCACUCUGCAAACACGA .....((((....))))(((((((((((.((((...(((.....)))....)))).)))))).)).......((((..((((....(((((.....)))))))))..))))....))).. ( -35.00) >psp.2973 5780483 120 - 5928787 CGUAACUUCGGGAGAAGGUGCGCCGGUGAGGGUGAAGCAUUUACUGCGUAAGCCCACGCCGGUCGAAGAUACCAGGCCGCUGCGACUGUUUAUUAAAAACACAGCACUCUGCAAACACGA .....((((....))))(((((((((((.((((...(((.....)))....)))).)))))).)).......((((..((((....(((((.....)))))))))..))))....))).. ( -40.30) >consensus CGUAACUUCGGGAGAAGGUGCGCCGGCGAGGGUGAAGGAUUUACUCCGUAAGCCCAUGCCGGUCGAAGAUACCAGGCCGCUGCGACUGUUUAUUAAAAACACAGCACUCUGCAAACACGA .....((((....))))(((((((((((.((((...(((.....)))....)))).)))))).)).......((((..((((....(((((.....)))))))))..))))....))).. (-38.58 = -37.05 + -1.53)
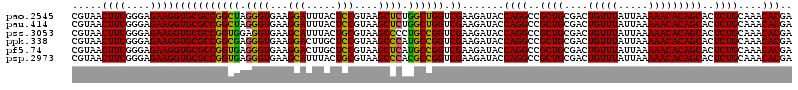
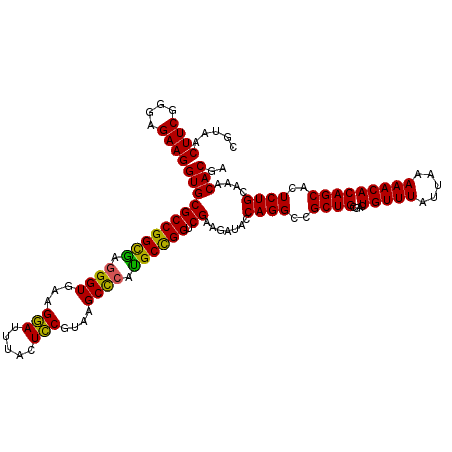
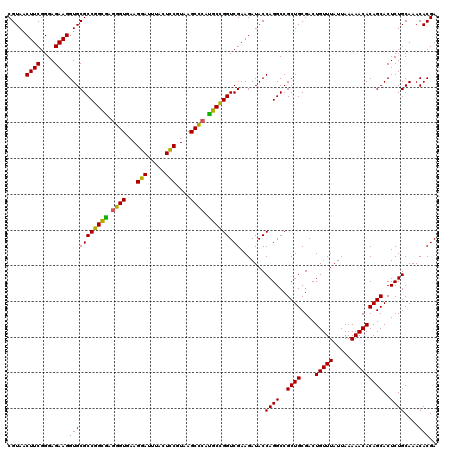
| Location | 4,789,970 – 4,790,090 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.56 |
| Mean single sequence MFE | -43.45 |
| Consensus MFE | -42.97 |
| Energy contribution | -41.45 |
| Covariance contribution | -1.52 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.33 |
| SVM RNA-class probability | 0.992537 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4789970 120 - 6264404 CUUGAGAGAACUCGGGUGAAGGAACUAGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGCUAGGGUGAAGGAUUUACUCCGUAAGCUCUGGCUGGUCGAAGAUACCAGGCCGC ((((((....))))))...........(((....(((((((....(....).....)))))((((((((((((...(((.....)))....)))))))))))).........)).))).. ( -44.80) >pau.414 5800801 120 + 6537648 CUUGAGAGAACUCGGGUGAAGGAACUAGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGCUAGGGUGAAGGAUUUACUCCGUAAGCUCUGGCUGGUCGAAGAUACCAGGCCGC ((((((....))))))...........(((....(((((((....(....).....)))))((((((((((((...(((.....)))....)))))))))))).........)).))).. ( -44.80) >pss.3053 5949357 120 - 6093698 CUUGAGAGAACUCGGGUGAAGGAACUAGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGUGGAGGUGAAGCAUUUACUGCGUAAGCCCCUGCCGGUCGAAGAUACCAGGCCGC ((((((....))))))...........(((....(((((((....(....).....)))))(((((..(.(((...(((.....)))....))).)..))))).........)).))).. ( -42.00) >ppk.338 5480258 120 + 6181863 CUUGAGAGAACUCGGGUGAAGGAACUAGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGCGAGGGUGAAGGACUUGCUCCGUAAGCCCAUGCCGGUCGAAGAUACCAGGCCGC ((((((....))))))...........(((....(((((((....(....).....)))))(((((((.((((...(((.....)))....)))).))))))).........)).))).. ( -45.00) >pf5.74 6948269 120 + 7074893 CUUGAGAGAACUCGGGUGAAGGAACUAGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGUGAGGGUGAAGGACUUGCUCCGUAAGCUCAUGCCGGUCGAAGAUACCAGGCCGC ((((((....))))))...........(((....(((((((....(....).....)))))(((((((.((((...(((.....)))....)))).))))))).........)).))).. ( -39.40) >psp.2973 5780523 120 - 5928787 CUUGAGAGAACUCGGGUGAAGGAACUAGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGUGAGGGUGAAGCAUUUACUGCGUAAGCCCACGCCGGUCGAAGAUACCAGGCCGC ((((((....))))))...........(((....(((((((....(....).....)))))(((((((.((((...(((.....)))....)))).))))))).........)).))).. ( -44.70) >consensus CUUGAGAGAACUCGGGUGAAGGAACUAGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGCGAGGGUGAAGGAUUUACUCCGUAAGCCCAUGCCGGUCGAAGAUACCAGGCCGC ((((((....))))))...........(((....(((((((....(....).....)))))(((((((.((((...(((.....)))....)))).))))))).........)).))).. (-42.97 = -41.45 + -1.52)

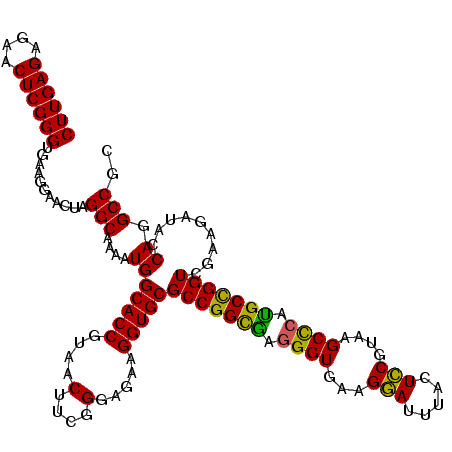
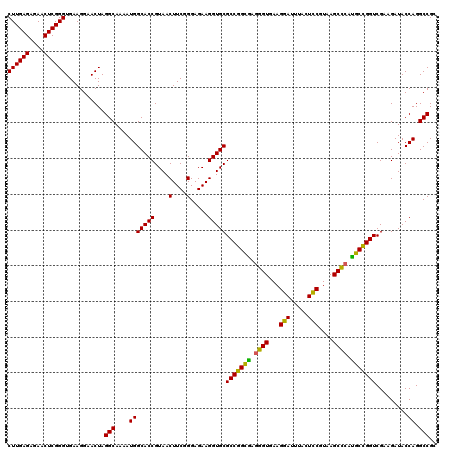
| Location | 4,790,010 – 4,790,130 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -30.90 |
| Consensus MFE | -28.99 |
| Energy contribution | -28.60 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.988802 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4790010 120 + 6264404 AUCCUUCACCCUAGCCGGCGCACCUUCUCCCGAAGUUACGGUGCCAUUUUGCCUAGUUCCUUCACCCGAGUUCUCUCAAGCGCCUUGGUAUUCUCUACCCGACCACCUGUGUCGGUUUGG .........((.(((((((((..........((((..(((((........)))..))..))))....(((....)))..)))))..((((.....)))).((((....).))))))).)) ( -31.90) >pau.414 5800841 120 - 6537648 AUCCUUCACCCUAGCCGGCGCACCUUCUCCCGAAGUUACGGUGCCAUUUUGCCUAGUUCCUUCACCCGAGUUCUCUCAAGCGCCUUGGUAUUCUCUACCCGACCACCUGUGUCGGUUUGG .........((.(((((((((..........((((..(((((........)))..))..))))....(((....)))..)))))..((((.....)))).((((....).))))))).)) ( -31.90) >pss.3053 5949397 120 + 6093698 AUGCUUCACCUCCACCGGCGCACCUUCUCCCGAAGUUACGGUGCCAUUUUGCCUAGUUCCUUCACCCGAGUUCUCUCAAGCGCCUUGGUAUUCUCUACCCAACCACCUGUGUCGGUUUGG ...((..(((..(((.(((((..........((((..(((((........)))..))..))))....(((....)))..)))))..((((.....)))).........)))..)))..)) ( -29.90) >pel.2704 4999928 120 + 5888780 GUGCUUCACCCUCACCGGCGCACCUUCUCCCGAAGUUACGGUGCCAUUUUGCCUAGUUCCUUCACCCGAGUUCUCUCAAGCGCCUUGGUAUUCUCUACCUAACCACCUGUGUCGGUUUGG ...((..(((..(((.(((((..........((((..(((((........)))..))..))))....(((....)))..)))))..((((.....)))).........)))..)))..)) ( -30.60) >ppk.338 5480298 120 - 6181863 GUCCUUCACCCUCGCCGGCGCACCUUCUCCCGAAGUUACGGUGCCAUUUUGCCUAGUUCCUUCACCCGAGUUCUCUCAAGCGCCUUGGUAUUCUCUACCUAACCACCUGUGUCGGUUUGG ..((...(((..(((.(((((..........((((..(((((........)))..))..))))....(((....)))..)))))..((((.....)))).........)))..)))..)) ( -30.90) >psp.2973 5780563 120 + 5928787 AUGCUUCACCCUCACCGGCGCACCUUCUCCCGAAGUUACGGUGCCAUUUUGCCUAGUUCCUUCACCCGAGUUCUCUCAAGCGCCUUGGUAUUCUCUACCCAACCACCUGUGUCGGUUUGG ...((..(((..(((.(((((..........((((..(((((........)))..))..))))....(((....)))..)))))..((((.....)))).........)))..)))..)) ( -30.20) >consensus AUCCUUCACCCUCACCGGCGCACCUUCUCCCGAAGUUACGGUGCCAUUUUGCCUAGUUCCUUCACCCGAGUUCUCUCAAGCGCCUUGGUAUUCUCUACCCAACCACCUGUGUCGGUUUGG .........((..((((((((..........((((..(((((........)))..))..))))....(((....)))..)))))..((((.....))))..............)))..)) (-28.99 = -28.60 + -0.39)

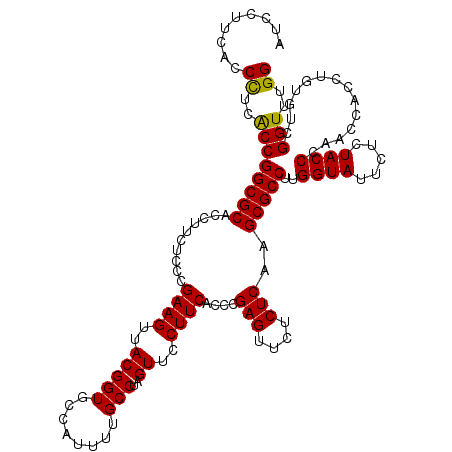
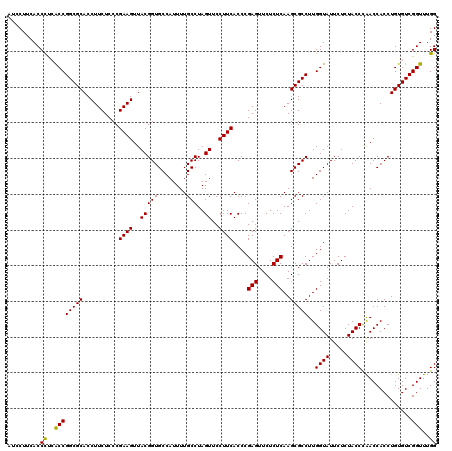
| Location | 4,790,010 – 4,790,130 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -38.58 |
| Consensus MFE | -32.47 |
| Energy contribution | -32.80 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.567965 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4790010 120 - 6264404 CCAAACCGACACAGGUGGUCGGGUAGAGAAUACCAAGGCGCUUGAGAGAACUCGGGUGAAGGAACUAGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGCUAGGGUGAAGGAU .....((..(((...(((((((((..((....((....((((((((....))))))))..))..))..))......(((((....(....).....))))).)))))))..)))..)).. ( -40.30) >pau.414 5800841 120 + 6537648 CCAAACCGACACAGGUGGUCGGGUAGAGAAUACCAAGGCGCUUGAGAGAACUCGGGUGAAGGAACUAGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGCUAGGGUGAAGGAU .....((..(((...(((((((((..((....((....((((((((....))))))))..))..))..))......(((((....(....).....))))).)))))))..)))..)).. ( -40.30) >pss.3053 5949397 120 - 6093698 CCAAACCGACACAGGUGGUUGGGUAGAGAAUACCAAGGCGCUUGAGAGAACUCGGGUGAAGGAACUAGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGUGGAGGUGAAGCAU ....(((..(((.(((((((.((((.....))))....((((((((....))))))))....))))..........(((((....(....).....)))))))).)))..)))....... ( -39.30) >pel.2704 4999928 120 - 5888780 CCAAACCGACACAGGUGGUUAGGUAGAGAAUACCAAGGCGCUUGAGAGAACUCGGGUGAAGGAACUAGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGUGAGGGUGAAGCAC ....(((..(((.(((((((.((((.....))))....((((((((....))))))))....))))..........(((((....(....).....)))))))).)))..)))....... ( -36.70) >ppk.338 5480298 120 + 6181863 CCAAACCGACACAGGUGGUUAGGUAGAGAAUACCAAGGCGCUUGAGAGAACUCGGGUGAAGGAACUAGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGCGAGGGUGAAGGAC ((..(((....((((((.((.((((.....)))).)).))))))......(((((((......))).(((......(((((....(....).....))))))))..)))))))...)).. ( -38.40) >psp.2973 5780563 120 - 5928787 CCAAACCGACACAGGUGGUUGGGUAGAGAAUACCAAGGCGCUUGAGAGAACUCGGGUGAAGGAACUAGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGUGAGGGUGAAGCAU ....(((..(((.(((((((.((((.....))))....((((((((....))))))))....))))..........(((((....(....).....)))))))).)))..)))....... ( -36.50) >consensus CCAAACCGACACAGGUGGUUGGGUAGAGAAUACCAAGGCGCUUGAGAGAACUCGGGUGAAGGAACUAGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGCGAGGGUGAAGCAU ((...((..((((((((.((.((((.....)))).)).)))))(((....)))..)))..)).....(((......(((((....(....).....)))))))))).............. (-32.47 = -32.80 + 0.33)

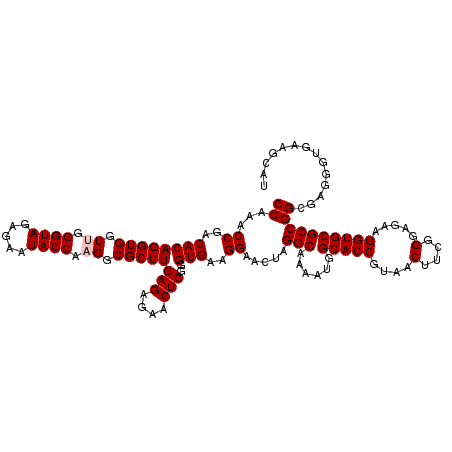
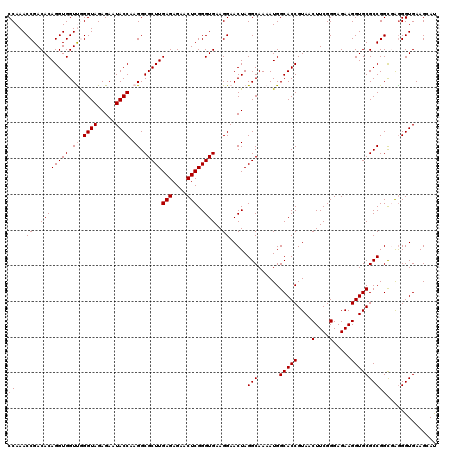
| Location | 4,790,130 – 4,790,250 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.26 |
| Mean single sequence MFE | -35.72 |
| Consensus MFE | -34.68 |
| Energy contribution | -32.52 |
| Covariance contribution | -2.16 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.02 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.777681 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4790130 120 - 6264404 UAGGUAAAUCCGGGGUUUCAAGGCCGAGAGCUGAUGACGAGUCGUCUUUUAGAUGACGAAGUGGUUGAUGCCAUGCUUCCAAGAAAAGCUUCUAAGCUUCAGGUAACCAGGAACCGUACC ..((((..(((..((((.(.((((..((((((........((((((.....))))))((((((((....)))..))))).......))).)))..))))...).)))).)))....)))) ( -35.90) >pss.3053 5949517 120 - 6093698 UAGGUAAAUCCGGGAUCUUAAGGCCGAGAGCUGAUGACGAGUGUUCUUUUAGAACACGAAGUGGUUGAUGCCAUGCUUCCAAGAAAAGCUUCUAAGCUUCAGGUAACCAGGAACCGUACC ..((((..(((((...(((.((((..((((((........((((((.....))))))((((((((....)))..))))).......))).)))..)))).)))...)).)))....)))) ( -36.00) >pel.2704 5000048 119 - 5888780 UAGGCAAAUCCGGGAUCUCAAGGCCGAGAGCUGAUGACGAGUGCUC-AUUAGAGCGCGAAGUGGUUGAUGCCAUGCUUCCAAGAAAAGCUCCUAAGCUUCAGAUAACUGGGAACCGUACC ..((....(((((.((((..((((..((((((........((((((-....))))))((((((((....)))..))))).......)))).))..)))).))))..)))))..))..... ( -34.90) >ppk.338 5480418 119 + 6181863 UAGGCAAAUCCGGGAUUUCAAGGCCGAGAGCUGAUGACGAGUUGCC-UUUAGGCGACGAAGUGGUUGAUACCAUGCUUCCAAGAAAAGCUCCUAAGCUUCAGAUAACUGGGAACCGUACC ..((....(((((...((..((((..((((((........((((((-....))))))((((((((....)))..))))).......)))).))..))))..))...)))))..))..... ( -33.70) >pf5.74 6948429 120 + 7074893 UAGGUAAAUCCGGGGUUCUAAGGCCGAGAGCUGAUGACGAGUUACCUUUUAGGUGACGAAGUGGUUGAUGCCAUGCUUCCAAGAAAAGCUUCUAAGCUUCAGAUAACUAGGAACCGUACC ..((((..(((.((..(((.((((..((((((........((((((.....))))))((((((((....)))..))))).......))).)))..)))).)))...)).)))....)))) ( -36.10) >pfo.66 6313299 119 + 6438405 UAGGUAAAUCCGGGGUUUCAAGGCCGAGAGCUGAUGACGAGUUGCC-UUUAGGCGACGAAGUGGUUGAUGCCAUGCUUCCAAGAAAAGCUUCUAAGCUUCAGAUAACCAGGAACCGUACC ..((((..(((..((((((.((((..((((((........((((((-....))))))((((((((....)))..))))).......))).)))..))))..)).)))).)))....)))) ( -37.70) >consensus UAGGUAAAUCCGGGAUUUCAAGGCCGAGAGCUGAUGACGAGUUGCC_UUUAGACGACGAAGUGGUUGAUGCCAUGCUUCCAAGAAAAGCUUCUAAGCUUCAGAUAACCAGGAACCGUACC ..((((..(((.((..((..((((..((((((........((((((.....))))))((((((((....)))..))))).......)))).))..))))..))...)).)))....)))) (-34.68 = -32.52 + -2.16)
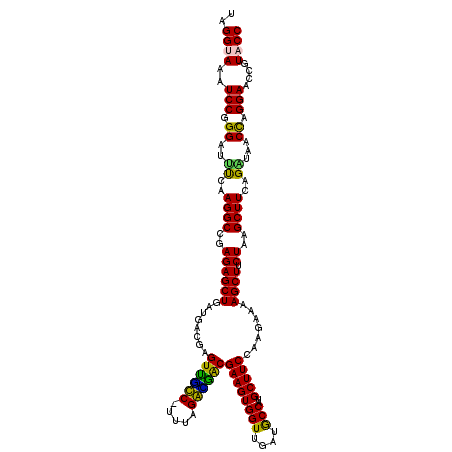
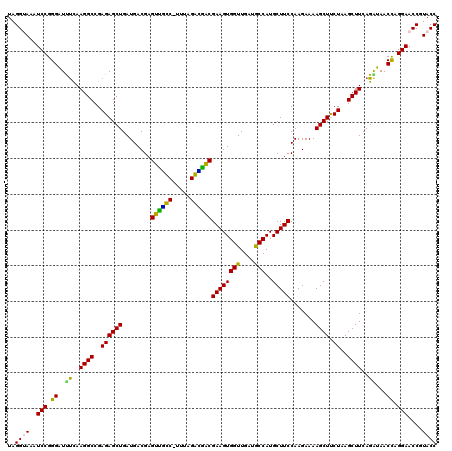
| Location | 4,790,170 – 4,790,290 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.88 |
| Mean single sequence MFE | -40.32 |
| Consensus MFE | -41.85 |
| Energy contribution | -38.67 |
| Covariance contribution | -3.19 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.58 |
| Structure conservation index | 1.04 |
| SVM decision value | 3.38 |
| SVM RNA-class probability | 0.999112 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4790170 120 - 6264404 GGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGAAAUCUUAGGUAAAUCCGGGGUUUCAAGGCCGAGAGCUGAUGACGAGUCGUCUUUUAGAUGACGAAGUGGUUGAUGCCAUGCUUCC ((((((..(..(....(((...(((....((((((((((((.((.....))))))))))..))))....)))...)))..((((((.....))))))...)..)..))))))........ ( -38.90) >pss.3053 5949557 120 - 6093698 GGCGUUGGUUGUCCAGGUUUAAGGUGGUAGGCUGAGAUCUUAGGUAAAUCCGGGAUCUUAAGGCCGAGAGCUGAUGACGAGUGUUCUUUUAGAACACGAAGUGGUUGAUGCCAUGCUUCC ((((((..(..(....(((...(((....((((((((((((.((.....))))))))))..))))....)))...)))..((((((.....))))))...)..)..))))))........ ( -40.40) >pel.2704 5000088 119 - 5888780 GGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGAGAUCUUAGGCAAAUCCGGGAUCUCAAGGCCGAGAGCUGAUGACGAGUGCUC-AUUAGAGCGCGAAGUGGUUGAUGCCAUGCUUCC ((((((..(..(....(((...(((....(((((((((((..((....))..)))))))..))))....)))...)))..((((((-....))))))...)..)..))))))........ ( -43.60) >ppk.338 5480458 119 + 6181863 GGCGUUGGUUGUCCAGGUUUAAGGUGGUAGGCUGAAAUCUUAGGCAAAUCCGGGAUUUCAAGGCCGAGAGCUGAUGACGAGUUGCC-UUUAGGCGACGAAGUGGUUGAUACCAUGCUUCC (((((.(((...(((.(((...(((....(((((((((((..((....))..)))))))..))))....)))...)))..((((((-....))))))....))).....))))))))... ( -39.60) >pf5.74 6948469 120 + 7074893 GGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGGAAUCUUAGGUAAAUCCGGGGUUCUAAGGCCGAGAGCUGAUGACGAGUUACCUUUUAGGUGACGAAGUGGUUGAUGCCAUGCUUCC ((((((..(..(....(((...(((....((((((((((((.((.....))))))))))..))))....)))...)))..((((((.....))))))...)..)..))))))........ ( -39.80) >psp.2973 5780723 119 - 5928787 GGCGUUGGUUGUCCAGGUUUAAGGUGGUAGGCUGAGAUCUUAGGUAAAUCCGGGAUUUCAAGGCCGAGAGCUGAUGACGAGUGUUC-UUUAGAACACGAAGUGGUUGAUGCCAUGCUUCC (((((..((((((..(((((....((((....(((((((((.((.....)))))))))))..)))).)))))...)))))((((((-....)))))).......)..)))))........ ( -39.60) >consensus GGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGAAAUCUUAGGUAAAUCCGGGAUUUCAAGGCCGAGAGCUGAUGACGAGUGGUC_UUUAGAACACGAAGUGGUUGAUGCCAUGCUUCC (((((.(((...(((.(((...(((....(((((((((((..((....))..)))))))..))))....)))...)))..((((((.....))))))....))).....))))))))... (-41.85 = -38.67 + -3.19)

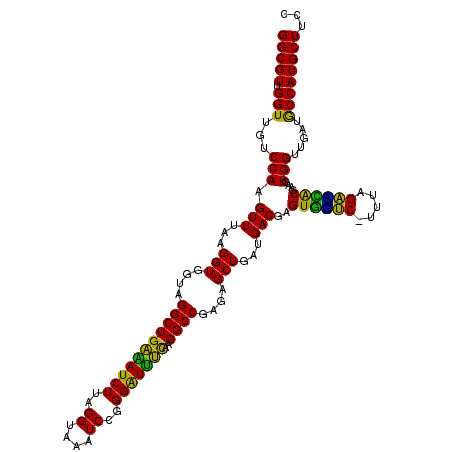
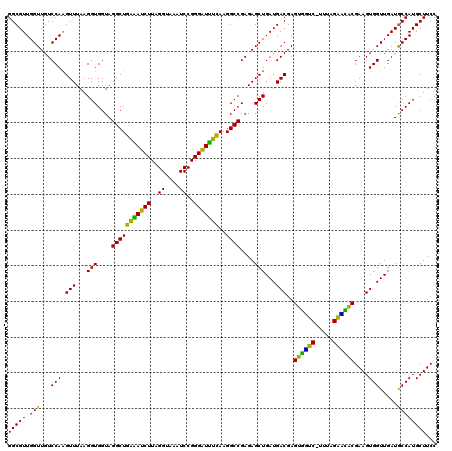
| Location | 4,790,210 – 4,790,330 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.11 |
| Mean single sequence MFE | -41.17 |
| Consensus MFE | -42.25 |
| Energy contribution | -40.83 |
| Covariance contribution | -1.41 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.22 |
| Structure conservation index | 1.03 |
| SVM decision value | 5.20 |
| SVM RNA-class probability | 0.999978 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4790210 120 - 6264404 GGUUACUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCUUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGAAAUCUUAGGUAAAUCCGGGGUUUCAAGGCCGAGAGCUGAUGACGA ................((((((..((.((((((....)))))).))..))))))..(((...(((....((((((((((((.((.....))))))))))..))))....)))...))).. ( -38.10) >pss.3053 5949597 120 - 6093698 GGUUAUUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCCUGGCGUUGGUUGUCCAGGUUUAAGGUGGUAGGCUGAGAUCUUAGGUAAAUCCGGGAUCUUAAGGCCGAGAGCUGAUGACGA .((((((((.......((((((..((.((((((....)))))).))..))))))...............((((((((((((.((.....))))))))))..))))....)).)))))).. ( -43.70) >pel.2704 5000127 120 - 5888780 AGUUAUUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCUUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGAGAUCUUAGGCAAAUCCGGGAUCUCAAGGCCGAGAGCUGAUGACGA .((((((((.......((((((..((.((((((....)))))).))..))))))...............(((((((((((..((....))..)))))))..))))....)).)))))).. ( -42.40) >ppk.338 5480497 120 + 6181863 AGUUAUUGCGAUGGAGGGACGGAGAAGGUUAGGCCAGCCUGGCGUUGGUUGUCCAGGUUUAAGGUGGUAGGCUGAAAUCUUAGGCAAAUCCGGGAUUUCAAGGCCGAGAGCUGAUGACGA .((((((((.......((((((..((.((((((....)))))).))..))))))...............(((((((((((..((....))..)))))))..))))....)).)))))).. ( -39.90) >pf5.74 6948509 120 + 7074893 AGUUAUUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCUUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGGAAUCUUAGGUAAAUCCGGGGUUCUAAGGCCGAGAGCUGAUGACGA .((((((((.......((((((..((.((((((....)))))).))..))))))...............((((((((((((.((.....))))))))))..))))....)).)))))).. ( -40.20) >psp.2973 5780762 120 - 5928787 GGUUAUUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCCUGGCGUUGGUUGUCCAGGUUUAAGGUGGUAGGCUGAGAUCUUAGGUAAAUCCGGGAUUUCAAGGCCGAGAGCUGAUGACGA .((((((((.......((((((..((.((((((....)))))).))..))))))...............((((((((((((.((.....))))))))))..))))....)).)))))).. ( -42.70) >consensus AGUUAUUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCCUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGAAAUCUUAGGUAAAUCCGGGAUUUCAAGGCCGAGAGCUGAUGACGA .((((((((.......((((((..((.((((((....)))))).))..))))))...............(((((((((((..((....))..)))))))..))))....)).)))))).. (-42.25 = -40.83 + -1.41)

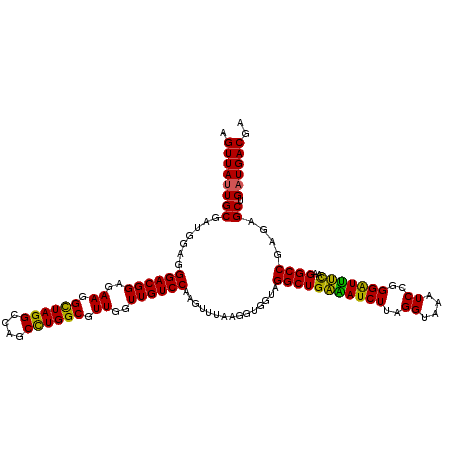

| Location | 4,790,250 – 4,790,370 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.28 |
| Mean single sequence MFE | -38.87 |
| Consensus MFE | -39.00 |
| Energy contribution | -37.50 |
| Covariance contribution | -1.50 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.70 |
| Structure conservation index | 1.00 |
| SVM decision value | 3.58 |
| SVM RNA-class probability | 0.999416 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4790250 120 - 6264404 GCGUAGUCGAUGGGAAACAGGUUAAUAUUCCUGUACUUCUGGUUACUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCUUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGAAAUCU .((((((.(((.(((((((((........)))))..)))).)))))))))..(((.((((((..((.((((((....)))))).))..)))))).((((((......))))))....))) ( -40.10) >pau.414 5801081 120 + 6537648 GCGUAGUCGAUGGGAAACAGGUUAAUAUUCCUGUACUUCUGGUUACUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCUUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGAAAUCU .((((((.(((.(((((((((........)))))..)))).)))))))))..(((.((((((..((.((((((....)))))).))..)))))).((((((......))))))....))) ( -40.10) >pel.2704 5000167 120 - 5888780 GCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUUCCAGUUAUUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCUUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGAGAUCU ...(((((..(((((.(((((........)))))..)))))..(((..(..((((.((((((..((.((((((....)))))).))..))))))...))))..)..))))))))...... ( -38.70) >ppk.338 5480537 120 + 6181863 GCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUUCCAGUUAUUGCGAUGGAGGGACGGAGAAGGUUAGGCCAGCCUGGCGUUGGUUGUCCAGGUUUAAGGUGGUAGGCUGAAAUCU .((((((...(((((.(((((........)))))..)))))...))))))......((((((..((.((((((....)))))).))..))))))((((((.((.(....).)).)))))) ( -39.40) >pf5.74 6948549 120 + 7074893 GCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUUCUAGUUAUUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCUUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGGAAUCU ........(((.....(((((........)))))...(((((((((..(..((((.((((((..((.((((((....)))))).))..))))))...))))..)..))).))))))))). ( -36.50) >psp.2973 5780802 120 - 5928787 GCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUUCUGGUUAUUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCCUGGCGUUGGUUGUCCAGGUUUAAGGUGGUAGGCUGAGAUCU .((((((...(((((.(((((........)))))..)))))...))))))......((((((..((.((((((....)))))).))..))))))((((((.((.(....).)).)))))) ( -38.40) >consensus GCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUUCUAGUUAUUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCUUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGAAAUCU .((((((...(((((.(((((........)))))..)))))...))))))..(((.((((((..((.((((((....)))))).))..)))))).((((((......))))))....))) (-39.00 = -37.50 + -1.50)
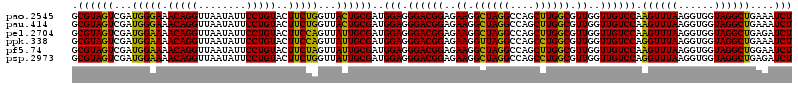
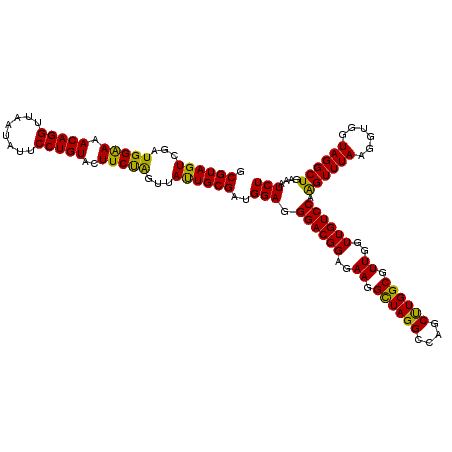
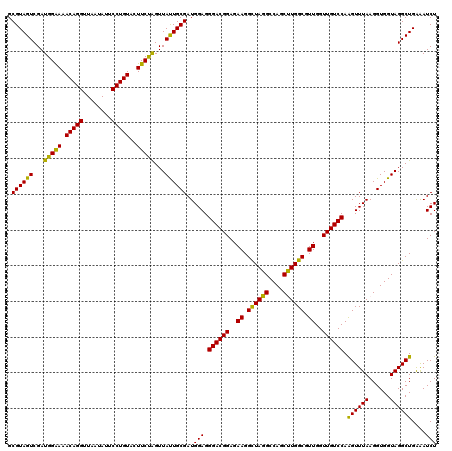
| Location | 4,790,290 – 4,790,410 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.72 |
| Mean single sequence MFE | -40.25 |
| Consensus MFE | -39.16 |
| Energy contribution | -38.08 |
| Covariance contribution | -1.08 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.09 |
| SVM RNA-class probability | 0.987701 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4790290 120 - 6264404 UCGACGCAGGGUUAGUCGGUUCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGGAAACAGGUUAAUAUUCCUGUACUUCUGGUUACUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCUU .....((.((.((((((.((((((....(...(((....)))(((((.(((.(((((((((........)))))..)))).)))))))))....))))))......)))))).)).)).. ( -41.30) >pau.414 5801121 120 + 6537648 UCGACGCAGGGUUAGUCGGUUCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGGAAACAGGUUAAUAUUCCUGUACUUCUGGUUACUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCUU .....((.((.((((((.((((((....(...(((....)))(((((.(((.(((((((((........)))))..)))).)))))))))....))))))......)))))).)).)).. ( -41.30) >pss.3053 5949677 120 - 6093698 UCGACGCAGGGUUAGUCGGUCCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUUCUGGUUAUUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCCU .....((.((.((((((.((((((....(...(((....)))(((((...(((((.(((((........)))))..)))))...))))))....))))))......)))))).)).)).. ( -38.70) >pel.2704 5000207 120 - 5888780 UCGACGCAGGGUUAGUCGGUCCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUUCCAGUUAUUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCUU .....((.((.((((((.((((((....(...(((....)))(((((...(((((.(((((........)))))..)))))...))))))....))))))......)))))).)).)).. ( -42.70) >ppk.338 5480577 120 + 6181863 UCGACGCAGGGUUAGUCGGUCCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUUCCAGUUAUUGCGAUGGAGGGACGGAGAAGGUUAGGCCAGCCU ........(((((..((.((((((....(...(((....)))(((((...(((((.(((((........)))))..)))))...))))))....)))))).))...((.....))))))) ( -40.50) >psp.2973 5780842 120 - 5928787 UCGACGCAGGGUUAGUCGGCCCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUUCUGGUUAUUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCCU ....(((.((((......)))).....)))((((((....((.(((((........(((((........))))).(((((.((....)).).))))..........))))).)))))))) ( -37.00) >consensus UCGACGCAGGGUUAGUCGGUCCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUUCUGGUUAUUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCCU .....((.((.((((((.((((((....(...(((....)))(((((...(((((.(((((........)))))..)))))...))))))....))))))......)))))).)).)).. (-39.16 = -38.08 + -1.08)

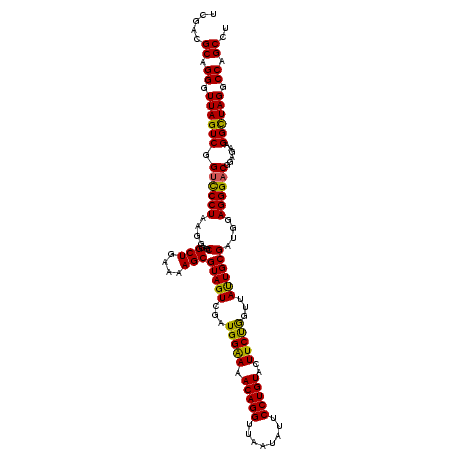
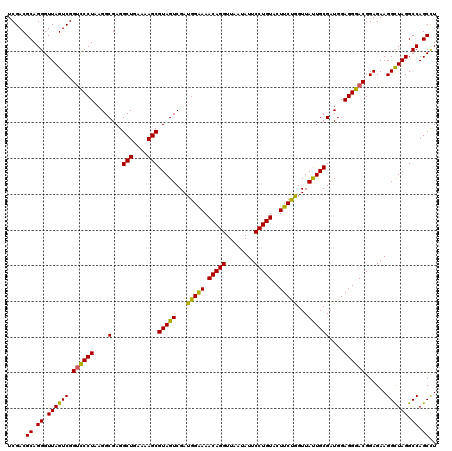
| Location | 4,790,330 – 4,790,450 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.06 |
| Mean single sequence MFE | -41.73 |
| Consensus MFE | -35.96 |
| Energy contribution | -36.60 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938805 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4790330 120 + 6264404 AGAAGUACAGGAAUAUUAACCUGUUUCCCAUCGACUACGCUUUUCAGCCUCGCCUUAGGAACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAACCCUUGGUCUUUCGGCGUGGGUG (((((((((((........)))))..............))))))((.((.((((..((((.((((.....((((((.((((....)))))))))).....))))))))..)))).)).)) ( -38.43) >pau.414 5801161 120 - 6537648 AGAAGUACAGGAAUAUUAACCUGUUUCCCAUCGACUACGCUUUUCAGCCUCGCCUUAGGAACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAACCCUUGGUCUUUCGGCGUGGGUG (((((((((((........)))))..............))))))((.((.((((..((((.((((.....((((((.((((....)))))))))).....))))))))..)))).)).)) ( -38.43) >pss.3053 5949717 120 + 6093698 AGAAGUACAGGAAUAUUAACCUGUUUUCCAUCGACUACGCUUUUCAGCCUCGCCUUAGGGACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGGGUG .((((.(((((........)))))))))................((.((.((((..(((((((((.....((((((.((((....)))))))))).....))))))))).)))).)).)) ( -42.60) >pel.2704 5000247 120 + 5888780 GGAAGUACAGGAAUAUUAACCUGUUUUCCAUCGACUACGCUUUUCAGCCUCGCCUUAGGGACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGCGAG (((((.(((((........)))))))))).(((.....(((....)))(.((((..(((((((((.....((((((.((((....)))))))))).....))))))))).)))).)))). ( -44.00) >ppk.338 5480617 120 - 6181863 GGAAGUACAGGAAUAUUAACCUGUUUUCCAUCGACUACGCUUUUCAGCCUCGCCUUAGGGACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGCGAG (((((.(((((........)))))))))).(((.....(((....)))(.((((..(((((((((.....((((((.((((....)))))))))).....))))))))).)))).)))). ( -44.00) >psp.2973 5780882 120 + 5928787 AGAAGUACAGGAAUAUUAACCUGUUUUCCAUCGACUACGCUUUUCAGCCUCGCCUUAGGGGCCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGGGUG .((((.(((((........)))))))))......(((((((.....(((((......))))).((((((.((((((.((((....)))))))))).....))))))....)))))))... ( -42.90) >consensus AGAAGUACAGGAAUAUUAACCUGUUUUCCAUCGACUACGCUUUUCAGCCUCGCCUUAGGGACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGGGUG (((((((((((........)))))..............))))))((.((.((((..(((((((((.....((((((.((((....)))))))))).....))))))))).)))).)).)) (-35.96 = -36.60 + 0.64)
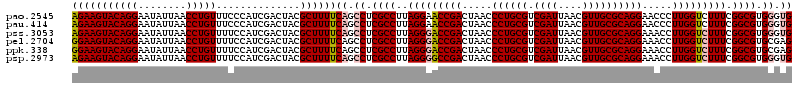
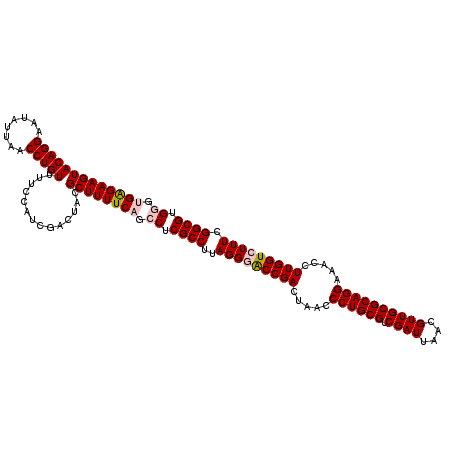
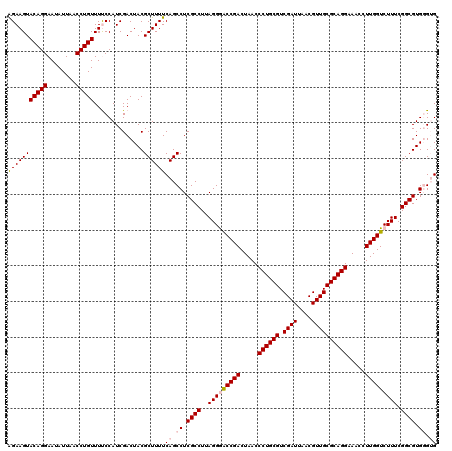
| Location | 4,790,330 – 4,790,450 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.06 |
| Mean single sequence MFE | -38.73 |
| Consensus MFE | -35.47 |
| Energy contribution | -34.97 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.860394 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4790330 120 - 6264404 CACCCACGCCGAAAGACCAAGGGUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUUCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGGAAACAGGUUAAUAUUCCUGUACUUCU ..((((((((....((((......(((((((...((.....)).)))))))......)))).....))))..(((....)))........))))..(((((........)))))...... ( -39.30) >pau.414 5801161 120 + 6537648 CACCCACGCCGAAAGACCAAGGGUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUUCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGGAAACAGGUUAAUAUUCCUGUACUUCU ..((((((((....((((......(((((((...((.....)).)))))))......)))).....))))..(((....)))........))))..(((((........)))))...... ( -39.30) >pss.3053 5949717 120 - 6093698 CACCCACGCCGAAAGACCAAGGUUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUCCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUUCU ...(((((((....((((......(((((((...((.....)).)))))))......)))).....))))..(((....)))........)))...(((((........)))))...... ( -37.60) >pel.2704 5000247 120 - 5888780 CUCGCACGCCGAAAGACCAAGGUUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUCCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUUCC .((((.((((....((((......(((((((...((.....)).)))))))......)))).....))))..(((....)))...).))).((((.(((((........)))))..)))) ( -39.40) >ppk.338 5480617 120 + 6181863 CUCGCACGCCGAAAGACCAAGGUUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUCCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUUCC .((((.((((....((((......(((((((...((.....)).)))))))......)))).....))))..(((....)))...).))).((((.(((((........)))))..)))) ( -39.40) >psp.2973 5780882 120 - 5928787 CACCCACGCCGAAAGACCAAGGUUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGCCCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUUCU .......(((((.((((....))))((((((...((.....)).)))))).....))))).((...(((.(.(((....))).).)))...))...(((((........)))))...... ( -37.40) >consensus CACCCACGCCGAAAGACCAAGGUUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUCCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUUCU ......((((....((((......(((((((...((.....)).)))))))......)))).....))))..(((....))).........((((.(((((........)))))..)))) (-35.47 = -34.97 + -0.50)
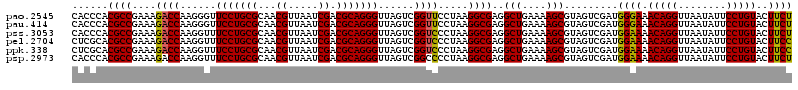
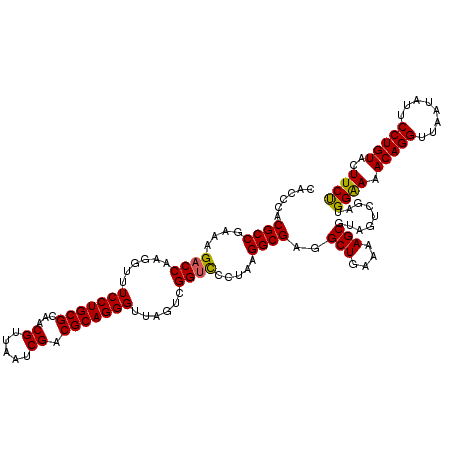
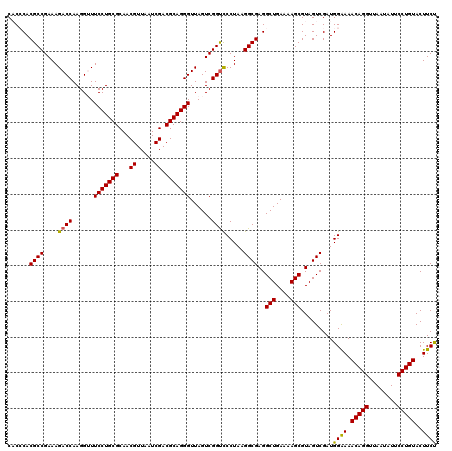
| Location | 4,790,370 – 4,790,490 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.06 |
| Mean single sequence MFE | -40.55 |
| Consensus MFE | -40.39 |
| Energy contribution | -39.70 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.52 |
| Structure conservation index | 1.00 |
| SVM decision value | 5.12 |
| SVM RNA-class probability | 0.999975 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4790370 120 + 6264404 UUUUCAGCCUCGCCUUAGGAACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAACCCUUGGUCUUUCGGCGUGGGUGUUUUUCACACCCAUUGUCGUUACUCAUGUCAGCAUUCGCA ......((..((((..((((.((((.....((((((.((((....)))))))))).....))))))))..))))(((((((.....))))))).....(((((....)).)))....)). ( -39.00) >pau.414 5801201 120 - 6537648 UUUUCAGCCUCGCCUUAGGAACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAACCCUUGGUCUUUCGGCGUGGGUGUUUUUCACACCCAUUGUCGUUACUCAUGUCAGCAUUCGCA ......((..((((..((((.((((.....((((((.((((....)))))))))).....))))))))..))))(((((((.....))))))).....(((((....)).)))....)). ( -39.00) >pel.2704 5000287 120 + 5888780 UUUUCAGCCUCGCCUUAGGGACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGCGAGUUUUUCACUCGCAUUGUCGUUACUCAUGUCAGCAUUCGCA ......((..((((..(((((((((.....((((((.((((....)))))))))).....))))))))).))))(((((((.....))))))).....(((((....)).)))....)). ( -40.90) >ppk.338 5480657 120 - 6181863 UUUUCAGCCUCGCCUUAGGGACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGCGAGUUUUUCACUCGCAUUGUCGUUACUCAUGUCAGCAUUCGCA ......((..((((..(((((((((.....((((((.((((....)))))))))).....))))))))).))))(((((((.....))))))).....(((((....)).)))....)). ( -40.90) >pfo.66 6313538 120 - 6438405 UUUUCAGCCUCGCCUUAGGGACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGGGUGUUUUUCACACCCAUUGUCGUUACUCAUGUCAGCAUUCGCA ......((..((((..(((((((((.....((((((.((((....)))))))))).....))))))))).))))(((((((.....))))))).....(((((....)).)))....)). ( -41.50) >psp.2973 5780922 120 + 5928787 UUUUCAGCCUCGCCUUAGGGGCCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGGGUGUUUUUCACACCCAUUGUCGUUACUCAUGUCAGCAUUCGCA ......(((((......))))).((((((.((((((.((((....)))))))))).....))))))...((((((((((((.....))))))).)))))............((....)). ( -42.00) >consensus UUUUCAGCCUCGCCUUAGGGACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGGGUGUUUUUCACACCCAUUGUCGUUACUCAUGUCAGCAUUCGCA ......((..((((..(((((((((.....((((((.((((....)))))))))).....))))))))).))))(((((((.....))))))).....(((((....)).)))....)). (-40.39 = -39.70 + -0.69)

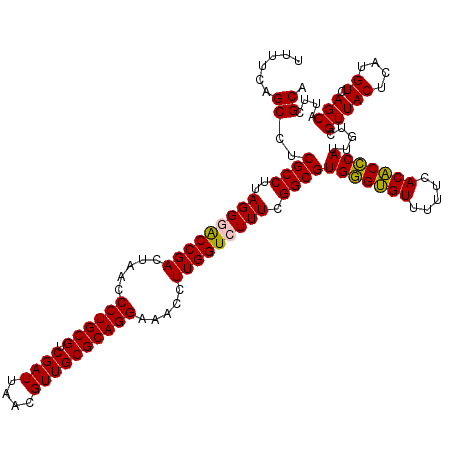
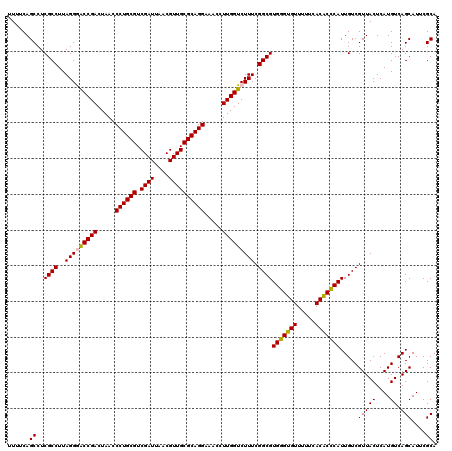
| Location | 4,790,370 – 4,790,490 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.06 |
| Mean single sequence MFE | -37.75 |
| Consensus MFE | -37.97 |
| Energy contribution | -37.03 |
| Covariance contribution | -0.94 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.50 |
| Structure conservation index | 1.01 |
| SVM decision value | 3.05 |
| SVM RNA-class probability | 0.998279 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4790370 120 - 6264404 UGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUGAAAAACACCCACGCCGAAAGACCAAGGGUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUUCCUAAGGCGAGGCUGAAAA .((...((((......)))).......(((((((.....)))))))((((....((((......(((((((...((.....)).)))))))......)))).....))))..))...... ( -36.80) >pau.414 5801201 120 + 6537648 UGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUGAAAAACACCCACGCCGAAAGACCAAGGGUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUUCCUAAGGCGAGGCUGAAAA .((...((((......)))).......(((((((.....)))))))((((....((((......(((((((...((.....)).)))))))......)))).....))))..))...... ( -36.80) >pel.2704 5000287 120 - 5888780 UGCGAAUGCUGACAUGAGUAACGACAAUGCGAGUGAAAAACUCGCACGCCGAAAGACCAAGGUUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUCCCUAAGGCGAGGCUGAAAA .((...((((......)))).......(((((((.....)))))))((((....((((......(((((((...((.....)).)))))))......)))).....))))..))...... ( -38.00) >ppk.338 5480657 120 + 6181863 UGCGAAUGCUGACAUGAGUAACGACAAUGCGAGUGAAAAACUCGCACGCCGAAAGACCAAGGUUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUCCCUAAGGCGAGGCUGAAAA .((...((((......)))).......(((((((.....)))))))((((....((((......(((((((...((.....)).)))))))......)))).....))))..))...... ( -38.00) >pfo.66 6313538 120 + 6438405 UGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUGAAAAACACCCACGCCGAAAGACCAAGGUUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUCCCUAAGGCGAGGCUGAAAA .((...((((......)))).......(((((((.....)))))))((((....((((......(((((((...((.....)).)))))))......)))).....))))..))...... ( -38.60) >psp.2973 5780922 120 - 5928787 UGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUGAAAAACACCCACGCCGAAAGACCAAGGUUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGCCCCUAAGGCGAGGCUGAAAA .(((..((((......)))).......(((((((.....))))))))))(....).........(((((((...((.....)).)))))))((((((.(((.....)))..))))))... ( -38.30) >consensus UGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUGAAAAACACCCACGCCGAAAGACCAAGGUUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUCCCUAAGGCGAGGCUGAAAA .((...((((......)))).......(((((((.....)))))))((((....((((......(((((((...((.....)).)))))))......)))).....))))..))...... (-37.97 = -37.03 + -0.94)
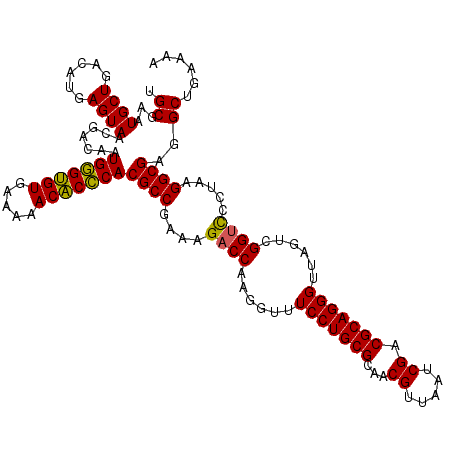
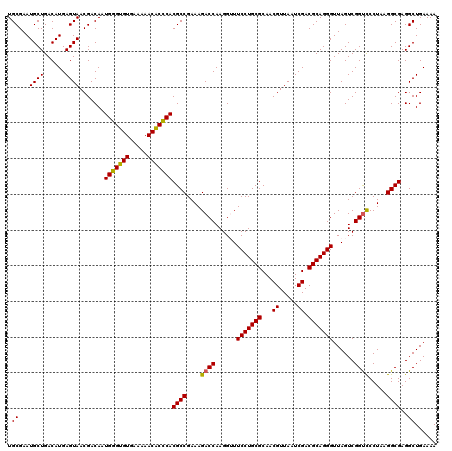
| Location | 4,790,410 – 4,790,530 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.78 |
| Mean single sequence MFE | -27.10 |
| Consensus MFE | -27.99 |
| Energy contribution | -27.10 |
| Covariance contribution | -0.89 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.04 |
| Structure conservation index | 1.03 |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.984573 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4790410 120 + 6264404 UUAACGUUGCGCAGGAACCCUUGGUCUUUCGGCGUGGGUGUUUUUCACACCCAUUGUCGUUACUCAUGUCAGCAUUCGCACUUCUGAUACCUCCAGCAAGCUUCUCAACUCACCUUCACA .....((((.(.(((.......(((....((((((((((((.....))))))).)))))..)))..((((((.(........)))))))))))))))....................... ( -27.30) >pau.414 5801241 120 - 6537648 UUAACGUUGCGCAGGAACCCUUGGUCUUUCGGCGUGGGUGUUUUUCACACCCAUUGUCGUUACUCAUGUCAGCAUUCGCACUUCUGAUACCUCCAGCAAGCUUCUCAACUCACCUUCACA .....((((.(.(((.......(((....((((((((((((.....))))))).)))))..)))..((((((.(........)))))))))))))))....................... ( -27.30) >pel.2704 5000327 120 + 5888780 UUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGCGAGUUUUUCACUCGCAUUGUCGUUACUCAUGUCAGCAUUCGCACUUCUGAUACCUCCAGCAAGCUUCUCAACUCACCUUCACA .....((((.(.(((.......(((....((((((((((((.....))))))).)))))..)))..((((((.(........)))))))))))))))....................... ( -26.70) >ppk.338 5480697 120 - 6181863 UUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGCGAGUUUUUCACUCGCAUUGUCGUUACUCAUGUCAGCAUUCGCACUUCUGAUACCUCCAGCAAGCUUCUCAACUCACCUUCACA .....((((.(.(((.......(((....((((((((((((.....))))))).)))))..)))..((((((.(........)))))))))))))))....................... ( -26.70) >pfo.66 6313578 120 - 6438405 UUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGGGUGUUUUUCACACCCAUUGUCGUUACUCAUGUCAGCAUUCGCACUUCUGAUACCUCCAGCAAGCUUCUCAACUCACCUUCACA .....((((.(.(((.......(((....((((((((((((.....))))))).)))))..)))..((((((.(........)))))))))))))))....................... ( -27.30) >psp.2973 5780962 120 + 5928787 UUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGGGUGUUUUUCACACCCAUUGUCGUUACUCAUGUCAGCAUUCGCACUUCUGAUACCUCCAGCAAGCUUCUCAACUCACCUUCACA .....((((.(.(((.......(((....((((((((((((.....))))))).)))))..)))..((((((.(........)))))))))))))))....................... ( -27.30) >consensus UUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGGGUGUUUUUCACACCCAUUGUCGUUACUCAUGUCAGCAUUCGCACUUCUGAUACCUCCAGCAAGCUUCUCAACUCACCUUCACA .....((((.(.(((.......(((....((((((((((((.....))))))).)))))..)))..((((((.(........)))))))))))))))....................... (-27.99 = -27.10 + -0.89)
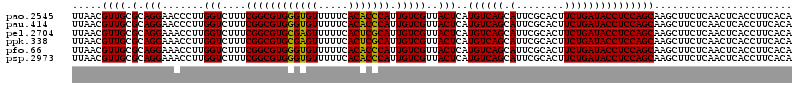
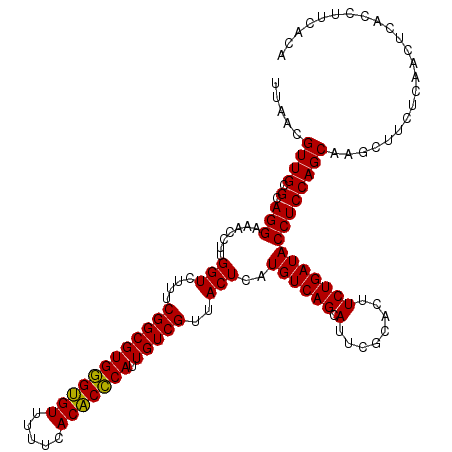
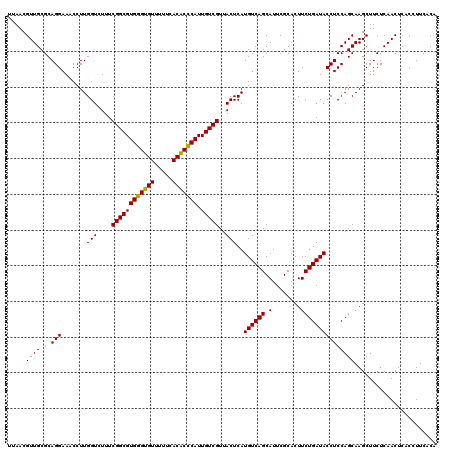
| Location | 4,790,410 – 4,790,530 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.78 |
| Mean single sequence MFE | -32.73 |
| Consensus MFE | -32.39 |
| Energy contribution | -31.50 |
| Covariance contribution | -0.89 |
| Combinations/Pair | 1.06 |
| Mean z-score | -0.86 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.852646 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4790410 120 - 6264404 UGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUGAAAAACACCCACGCCGAAAGACCAAGGGUUCCUGCGCAACGUUAA ((((..((..((((....))))..))..(((.(((......(((..((((......)))).......(((((((.....))))))))))(....)......))).))).))))....... ( -32.80) >pau.414 5801241 120 + 6537648 UGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUGAAAAACACCCACGCCGAAAGACCAAGGGUUCCUGCGCAACGUUAA ((((..((..((((....))))..))..(((.(((......(((..((((......)))).......(((((((.....))))))))))(....)......))).))).))))....... ( -32.80) >pel.2704 5000327 120 - 5888780 UGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGCGAGUGAAAAACUCGCACGCCGAAAGACCAAGGUUUCCUGCGCAACGUUAA ......((..((((....))))..))(..(((((..(.....)..)))))..).....(((((....(((((((.....))))))).((((..((((....))))..)).))..))))). ( -32.40) >ppk.338 5480697 120 + 6181863 UGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGCGAGUGAAAAACUCGCACGCCGAAAGACCAAGGUUUCCUGCGCAACGUUAA ......((..((((....))))..))(..(((((..(.....)..)))))..).....(((((....(((((((.....))))))).((((..((((....))))..)).))..))))). ( -32.40) >pfo.66 6313578 120 + 6438405 UGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUGAAAAACACCCACGCCGAAAGACCAAGGUUUCCUGCGCAACGUUAA ......((..((((....))))..))(..(((((..(.....)..)))))..).....(((((....(((((((.....))))))).((((..((((....))))..)).))..))))). ( -33.00) >psp.2973 5780962 120 - 5928787 UGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUGAAAAACACCCACGCCGAAAGACCAAGGUUUCCUGCGCAACGUUAA ......((..((((....))))..))(..(((((..(.....)..)))))..).....(((((....(((((((.....))))))).((((..((((....))))..)).))..))))). ( -33.00) >consensus UGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUGAAAAACACCCACGCCGAAAGACCAAGGUUUCCUGCGCAACGUUAA ((((..((((.((((....(((((.((..(((((..(.....)..)))))..)))))))..))))..(((((((.....))))))))))).........(((...))).))))....... (-32.39 = -31.50 + -0.89)

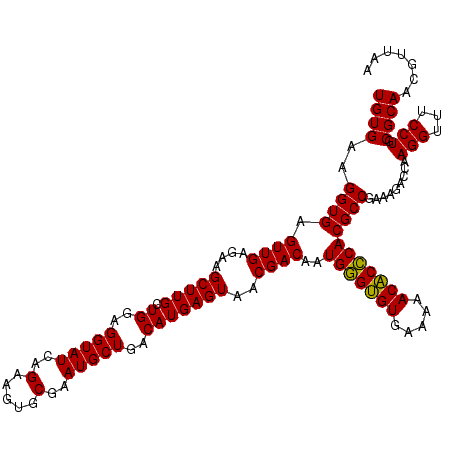
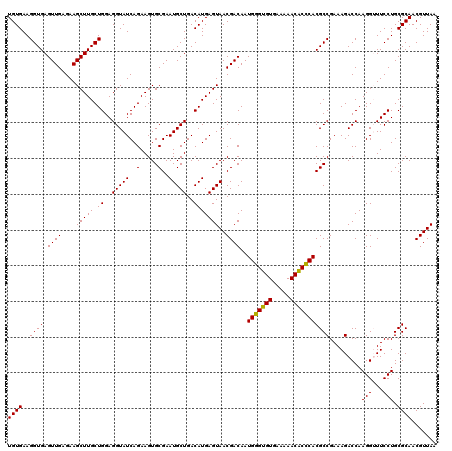
| Location | 4,790,450 – 4,790,568 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.11 |
| Mean single sequence MFE | -32.25 |
| Consensus MFE | -29.17 |
| Energy contribution | -29.00 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.940176 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4790450 118 - 6264404 UCACGUAA--GUGACGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUGAAAAA ((((....--))))..(((((.....((((((((...((((.(...((..((((....))))..))).))))..))))).)))...)))))((((..((.......))..))))...... ( -31.70) >pau.414 5801281 118 + 6537648 UCACGCAA--GUGACGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUGAAAAA ((((((..--.((.(((((((.....((((((((...((((.(...((..((((....))))..))).))))..))))).)))...)))))((....))..)).))....)))))).... ( -33.60) >pss.3053 5949837 120 - 6093698 UCAUCUUAUGAUGAUGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUGAAAAA (((((((((..(.((((((((.....((((((((...((((.(...((..((((....))))..))).))))..))))).)))...))))).))).)((....)).))))).)))).... ( -31.30) >pel.2704 5000367 119 - 5888780 UCACCUU-UGGUGAUGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGCGAGUGAAAAA ((((..(-((.(.((((((((.....((((((((...((((.(...((..((((....))))..))).))))..))))).)))...))))).))).).)))((......)).)))).... ( -32.90) >ppk.338 5480737 120 + 6181863 UCAUCUUAUGAUGAUGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGCGAGUGAAAAA (((((....)))))..(((((.....((((((((...((((.(...((..((((....))))..))).))))..))))).)))...))))).(((..(((.......)))..)))..... ( -32.70) >psp.2973 5781002 120 - 5928787 UCAUCUUUUGAUGAUGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUGAAAAA .(((((.(((.(.((((((((.....((((((((...((((.(...((..((((....))))..))).))))..))))).)))...))))).))).).......))).)))))....... ( -31.30) >consensus UCACCUUAUGAUGAUGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUGAAAAA (((((....))))).((((((.....((((((((...((((.(...((..((((....))))..))).))))..))))).)))...))))).)........................... (-29.17 = -29.00 + -0.17)

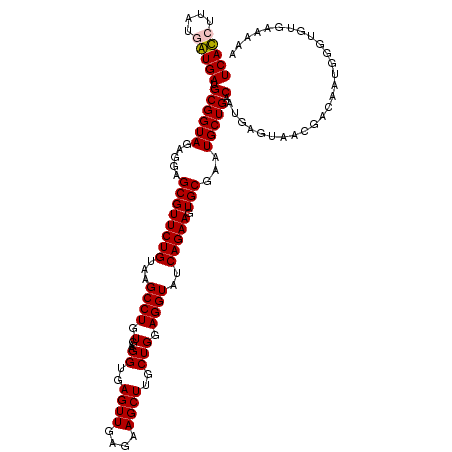
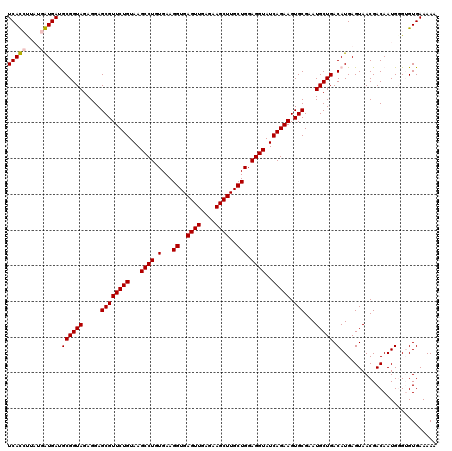
| Location | 4,790,490 – 4,790,608 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.87 |
| Mean single sequence MFE | -41.93 |
| Consensus MFE | -39.92 |
| Energy contribution | -39.03 |
| Covariance contribution | -0.89 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.95 |
| SVM decision value | 4.17 |
| SVM RNA-class probability | 0.999822 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4790490 118 - 6264404 GAUGUAACGGGGCUCAAACCACACACCGAAGCUGCGGGUGUCACGUAA--GUGACGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAG ...........((((..(((.....(((......)))(((((((....--))))))))))....)))).(((((...((((.(...((..((((....))))..))).))))..))))). ( -43.20) >pau.414 5801321 118 + 6537648 GAUGUAACGGGGCUCAAACCACACACCGAAGCUGCGGGUGUCACGCAA--GUGACGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAG ...........((((..(((.....(((......)))(((((((....--))))))))))....)))).(((((...((((.(...((..((((....))))..))).))))..))))). ( -44.20) >pel.2704 5000407 119 - 5888780 GAUGUAACGGGGCUCAAACCAUGCACCGAAGCUACGGGUAUCACCUU-UGGUGAUGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAG ...........((((..(((.....(((......)))((((((((..-.)))))))))))....)))).(((((...((((.(...((..((((....))))..))).))))..))))). ( -40.50) >ppk.338 5480777 120 + 6181863 GAUGUAACGGGGCUCAAACCAUGCACCGAAGCUACGGGUAUCAUCUUAUGAUGAUGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAG ...........((((..(((.....(((......)))((((((((....)))))))))))....)))).(((((...((((.(...((..((((....))))..))).))))..))))). ( -40.60) >pfo.66 6313658 118 + 6438405 GAUGUAACGGGGCUCAAACCAUACACCGAAGCUACGGGUAUCACGCAA--GUGAUGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAG ...........((((..(((.....(((......)))(((((((....--))))))))))....)))).(((((...((((.(...((..((((....))))..))).))))..))))). ( -42.50) >psp.2973 5781042 120 - 5928787 GAUGUAACGGGGCUCAAACCAUGCACCGAAGCUACGGGUAUCAUCUUUUGAUGAUGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAG ...........((((..(((.....(((......)))((((((((....)))))))))))....)))).(((((...((((.(...((..((((....))))..))).))))..))))). ( -40.60) >consensus GAUGUAACGGGGCUCAAACCAUACACCGAAGCUACGGGUAUCACCUAA__GUGAUGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAG ...........((((..(((.....(((......)))(((((((......))))))))))....)))).(((((...((((.(...((..((((....))))..))).))))..))))). (-39.92 = -39.03 + -0.89)

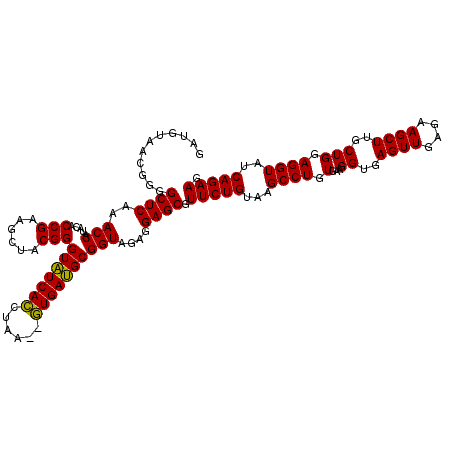
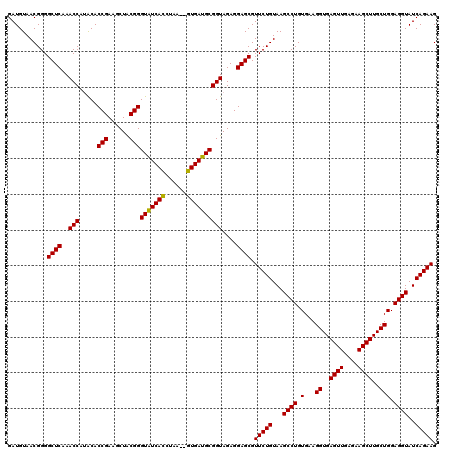
| Location | 4,790,608 – 4,790,728 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.67 |
| Mean single sequence MFE | -34.63 |
| Consensus MFE | -34.30 |
| Energy contribution | -34.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.04 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.942135 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4790608 120 - 6264404 AAAGUUGUGGUUAAGUGGUAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCAGCCACCCUUUAAAGAAAGCGUAAUAGCUCACUAGUCGAGUCGGCCUGCGCGGAA ......................((.((..((..(((((.(((.....(((((.(((((.......))))).)))))..((..(((......)))..)).))))))))..))..))))... ( -34.30) >pau.414 5801439 120 + 6537648 AAAGUUGUGGUUAAGUGGUAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCAGCCACCCUUUAAAGAAAGCGUAAUAGCUCACUAGUCGAGUCGGCCUGCGCGGAA ......................((.((..((..(((((.(((.....(((((.(((((.......))))).)))))..((..(((......)))..)).))))))))..))..))))... ( -34.30) >ppk.338 5480897 120 + 6181863 AAAGUCAUGGUUAAGUGGGAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCAGCCACCCUUUAAAGAAAGCGUAAUAGCUCACUAGUCGAGUCGGCCUGCGCGGAA ....((((......))))....((.((..((..(((((.(((.....(((((.(((((.......))))).)))))..((..(((......)))..)).))))))))..))..))))... ( -35.20) >pf5.74 6948907 120 + 7074893 AAAGUUAUGGUUAAGUGGGAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCAGCCACCCUUUAAAGAAAGCGUAAUAGCUCACUAGUCGAGUCGGCCUGCGCGGAA ..............((.(....).))(..((..(((((.(((.....(((((.(((((.......))))).)))))..((..(((......)))..)).))))))))..))..)...... ( -34.40) >pfo.66 6313776 120 + 6438405 AAAGUUAUGGUUAAGUGGGAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCAGCCACCCUUUAAAGAAAGCGUAAUAGCUCACUAGUCGAGUCGGCCUGCGCGGAA ..............((.(....).))(..((..(((((.(((.....(((((.(((((.......))))).)))))..((..(((......)))..)).))))))))..))..)...... ( -34.40) >psp.2973 5781162 120 - 5928787 AAAGUCAUGGUUAAGUGGGAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCAGCCACCCUUUAAAGAAAGCGUAAUAGCUCACUAGUCGAGUCGGCCUGCGCGGAA ....((((......))))....((.((..((..(((((.(((.....(((((.(((((.......))))).)))))..((..(((......)))..)).))))))))..))..))))... ( -35.20) >consensus AAAGUUAUGGUUAAGUGGGAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCAGCCACCCUUUAAAGAAAGCGUAAUAGCUCACUAGUCGAGUCGGCCUGCGCGGAA ......................((.((..((..(((((.(((.....(((((.(((((.......))))).)))))..((..(((......)))..)).))))))))..))..))))... (-34.30 = -34.30 + 0.00)

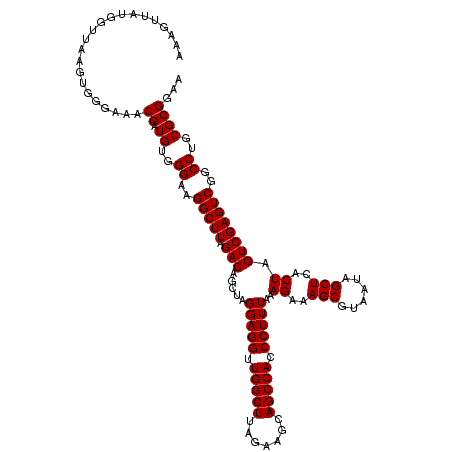
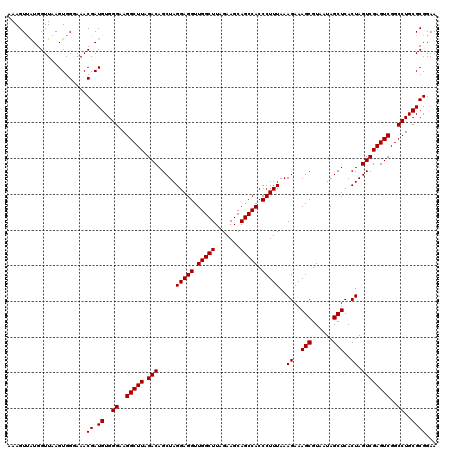
| Location | 4,790,648 – 4,790,768 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.22 |
| Mean single sequence MFE | -34.28 |
| Consensus MFE | -33.78 |
| Energy contribution | -33.34 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.99 |
| SVM decision value | 4.77 |
| SVM RNA-class probability | 0.999948 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4790648 120 + 6264404 CUUUAAAGGGUGGCUGCUUCUAAGCCAACCUCCUAGCUGUCUAAGCCUUCCCACAUCGUUUACCACUUAACCACAACUUUGGGACCUUAGCUGGCGGUCUGGGUUGUUUCCCUUUUCACG ....((((((.((((.......))))(((..((((((((((..(((..(((((....(((........)))........))))).....)))))))).)))))..))).))))))..... ( -32.60) >pau.414 5801479 120 - 6537648 CUUUAAAGGGUGGCUGCUUCUAAGCCAACCUCCUAGCUGUCUAAGCCUUCCCACAUCGUUUACCACUUAACCACAACUUUGGGACCUUAGCUGGCGGUCUGGGUUGUUUCCCUUUUCACG ....((((((.((((.......))))(((..((((((((((..(((..(((((....(((........)))........))))).....)))))))).)))))..))).))))))..... ( -32.60) >ppk.338 5480937 120 - 6181863 CUUUAAAGGGUGGCUGCUUCUAAGCCAACCUCCUAGCUGUCUAAGCCUUCCCACAUCGUUUCCCACUUAACCAUGACUUUGGGACCUUAGCUGACGGUCUGGGUUGUUUCCCUUUUCACG ....((((((.((((.......))))(((..((((((((((..(((..((((((((.(((........))).)))....))))).....)))))))).)))))..))).))))))..... ( -36.60) >pf5.74 6948947 120 - 7074893 CUUUAAAGGGUGGCUGCUUCUAAGCCAACCUCCUAGCUGUCUAAGCCUUCCCACAUCGUUUCCCACUUAACCAUAACUUUGGGACCUUAGCUGACGGUCUGGGUUGUUUCCCUUUUCACG ....((((((.((((.......))))(((..((((((((((..(((..(((((....(((..............)))..))))).....)))))))).)))))..))).))))))..... ( -33.64) >pfo.66 6313816 120 - 6438405 CUUUAAAGGGUGGCUGCUUCUAAGCCAACCUCCUAGCUGUCUAAGCCUUCCCACAUCGUUUCCCACUUAACCAUAACUUUGGGACCUUAGCUGACGGUCUGGGUUGUUUCCCUUUUCACG ....((((((.((((.......))))(((..((((((((((..(((..(((((....(((..............)))..))))).....)))))))).)))))..))).))))))..... ( -33.64) >psp.2973 5781202 120 + 5928787 CUUUAAAGGGUGGCUGCUUCUAAGCCAACCUCCUAGCUGUCUAAGCCUUCCCACAUCGUUUCCCACUUAACCAUGACUUUGGGACCUUAGCUGACGGUCUGGGUUGUUUCCCUUUUCACG ....((((((.((((.......))))(((..((((((((((..(((..((((((((.(((........))).)))....))))).....)))))))).)))))..))).))))))..... ( -36.60) >consensus CUUUAAAGGGUGGCUGCUUCUAAGCCAACCUCCUAGCUGUCUAAGCCUUCCCACAUCGUUUCCCACUUAACCAUAACUUUGGGACCUUAGCUGACGGUCUGGGUUGUUUCCCUUUUCACG ....((((((.((((.......))))(((..((((((((((..(((..(((((....(((..............)))..))))).....)))))))).)))))..))).))))))..... (-33.78 = -33.34 + -0.44)

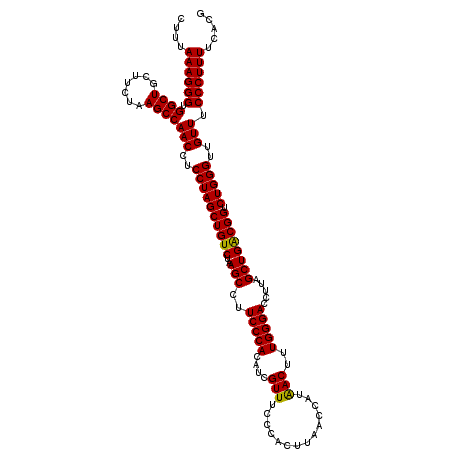
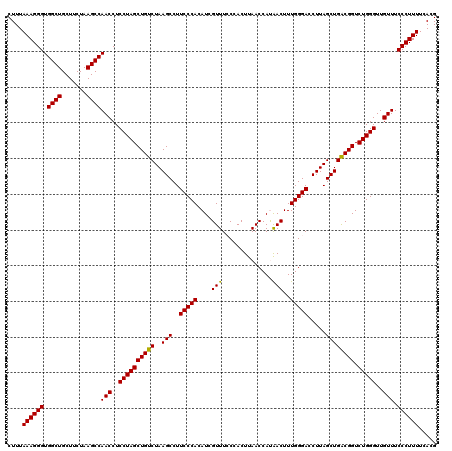
| Location | 4,790,648 – 4,790,768 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.22 |
| Mean single sequence MFE | -36.00 |
| Consensus MFE | -35.84 |
| Energy contribution | -35.73 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.68 |
| Structure conservation index | 1.00 |
| SVM decision value | 3.27 |
| SVM RNA-class probability | 0.998885 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4790648 120 - 6264404 CGUGAAAAGGGAAACAACCCAGACCGCCAGCUAAGGUCCCAAAGUUGUGGUUAAGUGGUAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCAGCCACCCUUUAAAG .....((((((......((((((((((.((((..(....)..))))))))))..((.....))....))))..((((....(((((....)))))(((....))))))).)))))).... ( -36.00) >pau.414 5801479 120 + 6537648 CGUGAAAAGGGAAACAACCCAGACCGCCAGCUAAGGUCCCAAAGUUGUGGUUAAGUGGUAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCAGCCACCCUUUAAAG .....((((((......((((((((((.((((..(....)..))))))))))..((.....))....))))..((((....(((((....)))))(((....))))))).)))))).... ( -36.00) >ppk.338 5480937 120 + 6181863 CGUGAAAAGGGAAACAACCCAGACCGUCAGCUAAGGUCCCAAAGUCAUGGUUAAGUGGGAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCAGCCACCCUUUAAAG .....((((((......((((((((((..(((..(....)..))).))))))..((.(....).)).))))..((((....(((((....)))))(((....))))))).)))))).... ( -35.10) >pf5.74 6948947 120 + 7074893 CGUGAAAAGGGAAACAACCCAGACCGUCAGCUAAGGUCCCAAAGUUAUGGUUAAGUGGGAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCAGCCACCCUUUAAAG .....((((((......((((((((((.((((..(....)..))))))))))..((.(....).)).))))..((((....(((((....)))))(((....))))))).)))))).... ( -36.90) >pfo.66 6313816 120 + 6438405 CGUGAAAAGGGAAACAACCCAGACCGUCAGCUAAGGUCCCAAAGUUAUGGUUAAGUGGGAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCAGCCACCCUUUAAAG .....((((((......((((((((((.((((..(....)..))))))))))..((.(....).)).))))..((((....(((((....)))))(((....))))))).)))))).... ( -36.90) >psp.2973 5781202 120 - 5928787 CGUGAAAAGGGAAACAACCCAGACCGUCAGCUAAGGUCCCAAAGUCAUGGUUAAGUGGGAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCAGCCACCCUUUAAAG .....((((((......((((((((((..(((..(....)..))).))))))..((.(....).)).))))..((((....(((((....)))))(((....))))))).)))))).... ( -35.10) >consensus CGUGAAAAGGGAAACAACCCAGACCGUCAGCUAAGGUCCCAAAGUUAUGGUUAAGUGGGAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCAGCCACCCUUUAAAG .....((((((......((((((((((.((((..(....)..))))))))))..((.(....).)).))))..((((....(((((....)))))(((....))))))).)))))).... (-35.84 = -35.73 + -0.11)
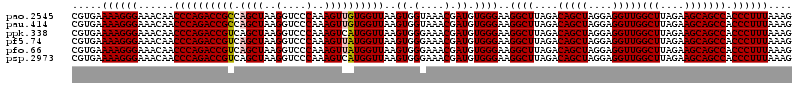
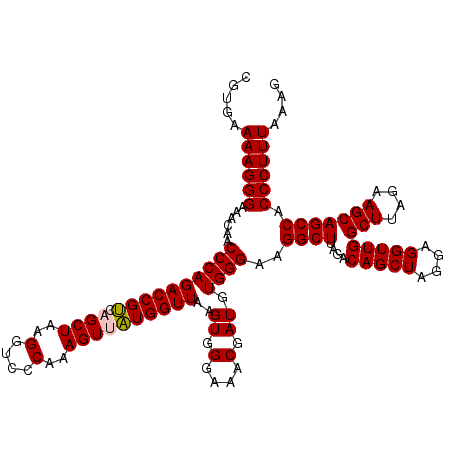
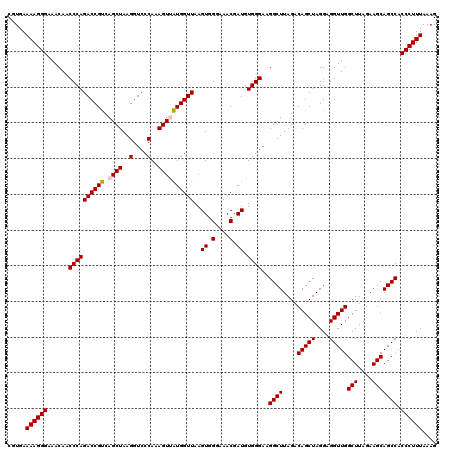
| Location | 4,790,688 – 4,790,808 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.22 |
| Mean single sequence MFE | -40.91 |
| Consensus MFE | -39.34 |
| Energy contribution | -39.23 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.73 |
| SVM RNA-class probability | 0.996690 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4790688 120 - 6264404 UGCCGAGCAUGGGAGACACACGGCGGGUGCUAACGUCCGUCGUGAAAAGGGAAACAACCCAGACCGCCAGCUAAGGUCCCAAAGUUGUGGUUAAGUGGUAAACGAUGUGGGAAGGCUUAG .(((.(((.(((......((((((((..(....)..))))))))....(((......)))......))))))....(((((..(((((.............))))).))))).))).... ( -41.32) >pau.414 5801519 120 + 6537648 UGCCGAGCAUGGGAGACACACGGCGGGUGCUAACGUCCGUCGUGAAAAGGGAAACAACCCAGACCGCCAGCUAAGGUCCCAAAGUUGUGGUUAAGUGGUAAACGAUGUGGGAAGGCUUAG .(((.(((.(((......((((((((..(....)..))))))))....(((......)))......))))))....(((((..(((((.............))))).))))).))).... ( -41.32) >ppk.338 5480977 120 + 6181863 UGCCGAGCAUGGGAGACACACGGCGGGUGCUAACGUCCGUCGUGAAAAGGGAAACAACCCAGACCGUCAGCUAAGGUCCCAAAGUCAUGGUUAAGUGGGAAACGAUGUGGGAAGGCUUAG .(((.(((((((......((((((((..(....)..))))))))....(((......)))...))))..)))....(((((....(((.(((........))).)))))))).))).... ( -41.10) >pf5.74 6948987 120 + 7074893 UGCCGAGCAUGGGAGACACACGGCGGGUGCUAACGUCCGUCGUGAAAAGGGAAACAACCCAGACCGUCAGCUAAGGUCCCAAAGUUAUGGUUAAGUGGGAAACGAUGUGGGAAGGCUUAG .(((..((((.(......((((((((..(....)..)))))))).....(....)..((((((((((.((((..(....)..))))))))))...))))...).)))).....))).... ( -40.30) >pfo.66 6313856 120 + 6438405 UGCCGAGCAUGGGAGACACACGGCGGGUGCUAACGUCCGUCGUGAAAAGGGAAACAACCCAGACCGUCAGCUAAGGUCCCAAAGUUAUGGUUAAGUGGGAAACGAUGUGGGAAGGCUUAG .(((..((((.(......((((((((..(....)..)))))))).....(....)..((((((((((.((((..(....)..))))))))))...))))...).)))).....))).... ( -40.30) >psp.2973 5781242 120 - 5928787 UGCCGAGCAUGGGAGACACACGGCGGGUGCUAACGUCCGUCGUGAAAAGGGAAACAACCCAGACCGUCAGCUAAGGUCCCAAAGUCAUGGUUAAGUGGGAAACGAUGUGGGAAGGCUUAG .(((.(((((((......((((((((..(....)..))))))))....(((......)))...))))..)))....(((((....(((.(((........))).)))))))).))).... ( -41.10) >consensus UGCCGAGCAUGGGAGACACACGGCGGGUGCUAACGUCCGUCGUGAAAAGGGAAACAACCCAGACCGUCAGCUAAGGUCCCAAAGUUAUGGUUAAGUGGGAAACGAUGUGGGAAGGCUUAG .(((.(((((((......((((((((..(....)..))))))))....(((......)))...))))..)))....(((((....(((.(((........))).)))))))).))).... (-39.34 = -39.23 + -0.11)

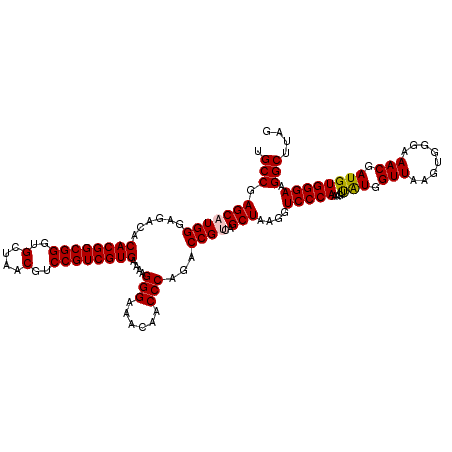

| Location | 4,790,728 – 4,790,848 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.67 |
| Mean single sequence MFE | -34.51 |
| Consensus MFE | -33.35 |
| Energy contribution | -33.69 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.82 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.683483 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4790728 120 - 6264404 CCGACUUACCAAACCGAUGCAAACUCCGAAUACCCAGAAGUGCCGAGCAUGGGAGACACACGGCGGGUGCUAACGUCCGUCGUGAAAAGGGAAACAACCCAGACCGCCAGCUAAGGUCCC ..(((((.......((..(((...((.(......).))..)))))(((.(((......((((((((..(....)..))))))))....(((......)))......)))))).))))).. ( -33.10) >pau.414 5801559 120 + 6537648 CCGACUUACCAAACCGAUGCAAACUCCGAAUACCCAGAAGUGCCGAGCAUGGGAGACACACGGCGGGUGCUAACGUCCGUCGUGAAAAGGGAAACAACCCAGACCGCCAGCUAAGGUCCC ..(((((.......((..(((...((.(......).))..)))))(((.(((......((((((((..(....)..))))))))....(((......)))......)))))).))))).. ( -33.10) >ppk.338 5481017 120 + 6181863 CCGACUUACCAAACCGAUGCAAACUCCGAAUACCUACAAGUGCCGAGCAUGGGAGACACACGGCGGGUGCUAACGUCCGUCGUGAAAAGGGAAACAACCCAGACCGUCAGCUAAGGUCCC ..(((((......((.((((..(((.............))).....)))).)).(((.((((((((..(....)..))))))))....(((......))).....))).....))))).. ( -35.22) >pf5.74 6949027 120 + 7074893 CCGACUUACCAAACCGAUGCAAACUCCGAAUACCUACAAGUGCCGAGCAUGGGAGACACACGGCGGGUGCUAACGUCCGUCGUGAAAAGGGAAACAACCCAGACCGUCAGCUAAGGUCCC ..(((((......((.((((..(((.............))).....)))).)).(((.((((((((..(....)..))))))))....(((......))).....))).....))))).. ( -35.22) >pfo.66 6313896 120 + 6438405 CCGACUUACCAAACCGAUGCAAACUCCGAAUACCUACAAGUGCCGAGCAUGGGAGACACACGGCGGGUGCUAACGUCCGUCGUGAAAAGGGAAACAACCCAGACCGUCAGCUAAGGUCCC ..(((((......((.((((..(((.............))).....)))).)).(((.((((((((..(....)..))))))))....(((......))).....))).....))))).. ( -35.22) >psp.2973 5781282 120 - 5928787 CCGACUUACCAAACCGAUGCAAACUCCGAAUACCUACAAGUGCCGAGCAUGGGAGACACACGGCGGGUGCUAACGUCCGUCGUGAAAAGGGAAACAACCCAGACCGUCAGCUAAGGUCCC ..(((((......((.((((..(((.............))).....)))).)).(((.((((((((..(....)..))))))))....(((......))).....))).....))))).. ( -35.22) >consensus CCGACUUACCAAACCGAUGCAAACUCCGAAUACCUACAAGUGCCGAGCAUGGGAGACACACGGCGGGUGCUAACGUCCGUCGUGAAAAGGGAAACAACCCAGACCGUCAGCUAAGGUCCC ..(((((......((.((((..(((.............))).....)))).)).(((.((((((((..(....)..))))))))....(((......))).....))).....))))).. (-33.35 = -33.69 + 0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:09:17 2007