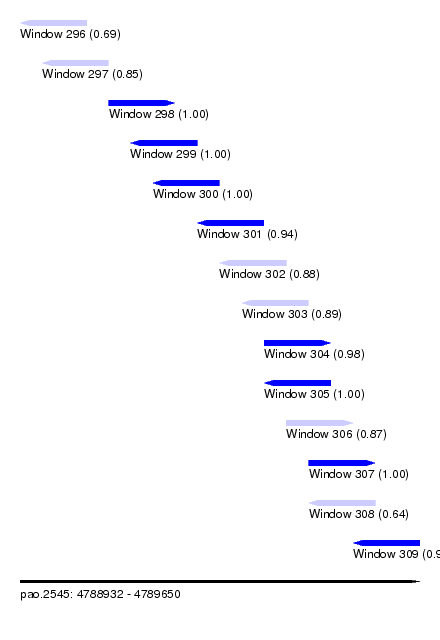
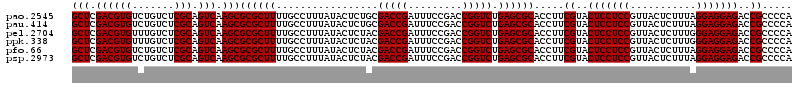
| Sequence ID | pao.2545 |
|---|---|
| Location | 4,788,932 – 4,789,650 |
| Length | 718 |
| Max. P | 0.999842 |

| Location | 4,788,932 – 4,789,052 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.06 |
| Mean single sequence MFE | -37.50 |
| Consensus MFE | -37.07 |
| Energy contribution | -35.68 |
| Covariance contribution | -1.39 |
| Combinations/Pair | 1.12 |
| Mean z-score | -0.72 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.694273 |
| Prediction | RNA |
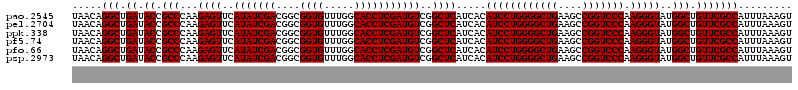
Download alignment: ClustalW | MAF
>pao.2545 4788932 120 - 6264404 UUCCGGUUGUCACGCCAGUGGCAUUGCCGGGUAGCUAUGUUCGGAAAAGAUAACCGCUGAAAGCAUCUAAGCGGGAAACUUGCCUCAAGAUGAGAUCUCACUGGGAACUUGAUUCCCCUG ...((((((((..(((...)))....(((..((....))..)))....))))))))..((...(((((..((((....).)))....)))))...)).....(((((.....)))))... ( -35.30) >pau.414 5799761 120 + 6537648 UUCCGGUUGUCACGCCAGUGGCAUUGCCGGGUAGCUAUGUUCGGAAAAGAUAACCGCUGAAAGCAUCUAAGCGGGAAACUUGCCUCAAGAUGAGAUCUCACUGGGAACUUGAUUCCCCUG ...((((((((..(((...)))....(((..((....))..)))....))))))))..((...(((((..((((....).)))....)))))...)).....(((((.....)))))... ( -35.30) >pel.2704 4998848 120 - 5888780 UUCCGGUUGUCACGCCAGUGGCAUUGCCGGGUAGCUAUGUUCGGAAGAGAUAACCGCUGAAAGCAUCUAAGCGGGAAACUUGCCUCAAGAUGAGAUCUCACUGGAGCCUUGAGCUCCCUG ....(((((((((....))))))..)))(((.((((..((((.(..(((((..(((((...........))))).........(((.....))))))))..).))))....))))))).. ( -41.20) >ppk.338 5479218 120 + 6181863 UUCCGGUUGUCACGCCAGUGGCAUUGCCGGGUAGCUAUGUUCGGAAGAGAUAACCGCUGAAAGCAUCUAAGCGGGAAACUUGCCUCAAGAUGAGAUCUCACUGGGAUCUUGAAUCCCCUA .((((((((((((....))))))..))))))((((....(((....)))......))))...((......))((((..((((...))))..(((((((.....)))))))...))))... ( -39.40) >pfo.66 6312099 120 + 6438405 UUCCGGUUGUCACGCCAGUGGCAUUGCCGGGUAGCUAUGUUCGGGAAAGAUAACCGCUGAAAGCAUCUAAGCGGGAAACUUGCCUCAAGAUGAGAUCUCACUGGAACCUUGAGUUCCCUG ....(((((((((....))))))..)))(((.((((..((((.((..((((..(((((...........))))).........(((.....)))))))..)).))))....))))))).. ( -38.70) >psp.2973 5779483 120 - 5928787 UUCCGGUUGUCACGCCAGUGGCAUUGCCGGGUAGCUAUGUUCGGAAAAGAUAACCGCUGAAAGCAUCUAAGCGGGAAACUUGCCUCAAGAUGAGAUCUCACUGGAACCUUGAGUUCCCUA ...((((((((..(((...)))....(((..((....))..)))....))))))))..((...(((((..((((....).)))....)))))...)).....(((((.....)))))... ( -35.10) >consensus UUCCGGUUGUCACGCCAGUGGCAUUGCCGGGUAGCUAUGUUCGGAAAAGAUAACCGCUGAAAGCAUCUAAGCGGGAAACUUGCCUCAAGAUGAGAUCUCACUGGAACCUUGAGUCCCCUG ...((((((((..(((...)))....(((..((....))..)))....))))))))..((...(((((..((((....).)))....)))))...)).....(((((.....)))))... (-37.07 = -35.68 + -1.39)
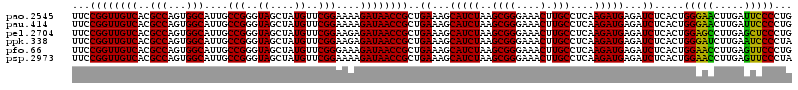
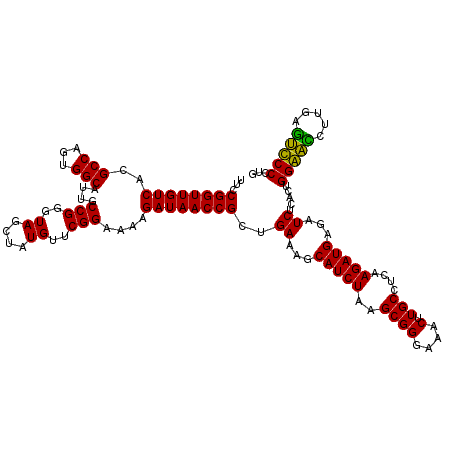
| Location | 4,788,972 – 4,789,091 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.00 |
| Mean single sequence MFE | -41.28 |
| Consensus MFE | -40.55 |
| Energy contribution | -40.72 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.91 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.846093 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4788972 119 - 6264404 CUCCUAGUACGAGAGGACCGGAGUGGACGAACCUCUGG-GUUCCGGUUGUCACGCCAGUGGCAUUGCCGGGUAGCUAUGUUCGGAAAAGAUAACCGCUGAAAGCAUCUAAGCGGGAAACU .((((.((...((((((((((((.(......)))))))-..)))(((((((..(((...)))....(((..((....))..)))....)))))))((.....)).)))..)))))).... ( -39.50) >pel.2704 4998888 120 - 5888780 CUCCUAGUACGAGAGGACCGGAGUGGACGAACCUCUGGUGUUCCGGUUGUCACGCCAGUGGCAUUGCCGGGUAGCUAUGUUCGGAAGAGAUAACCGCUGAAAGCAUCUAAGCGGGAAACU .(((((((..(.(((.(((((((.(......)))))))).))))(((((((..(((...)))....(((..((....))..)))....))))))))))....((......)))))).... ( -41.40) >ppk.338 5479258 120 + 6181863 CUCCUAGUACGAGAGGACCGGAGUGGACGAACCUCUGGUGUUCCGGUUGUCACGCCAGUGGCAUUGCCGGGUAGCUAUGUUCGGAAGAGAUAACCGCUGAAAGCAUCUAAGCGGGAAACU .(((((((..(.(((.(((((((.(......)))))))).))))(((((((..(((...)))....(((..((....))..)))....))))))))))....((......)))))).... ( -41.40) >pf5.74 6947269 120 + 7074893 CUCCUAGUACGAGAGGACCGGAGUGGACGAACCUCUGGUGUUCCGGUUGUCACGCCAGUGGCAUUGCCGGGUAGCUAUGUUCGGAAAAGAUAACCGCUGAAAGCAUCUAAGCGGGAAACU .(((((((..(.(((.(((((((.(......)))))))).))))(((((((..(((...)))....(((..((....))..)))....))))))))))....((......)))))).... ( -41.40) >pfo.66 6312139 120 + 6438405 CUCCUAGUACGAGAGGACCGGAGUGGACGAACCUCUGGUGUUCCGGUUGUCACGCCAGUGGCAUUGCCGGGUAGCUAUGUUCGGGAAAGAUAACCGCUGAAAGCAUCUAAGCGGGAAACU .(((((((..(.(((.(((((((.(......)))))))).))))(((((((..(((...))).((.(((..((....))..))).)).))))))))))....((......)))))).... ( -42.60) >psp.2973 5779523 120 - 5928787 CUCCUAGUACGAGAGGACCGGAGUGGACGAACCUCUGGUGUUCCGGUUGUCACGCCAGUGGCAUUGCCGGGUAGCUAUGUUCGGAAAAGAUAACCGCUGAAAGCAUCUAAGCGGGAAACU .(((((((..(.(((.(((((((.(......)))))))).))))(((((((..(((...)))....(((..((....))..)))....))))))))))....((......)))))).... ( -41.40) >consensus CUCCUAGUACGAGAGGACCGGAGUGGACGAACCUCUGGUGUUCCGGUUGUCACGCCAGUGGCAUUGCCGGGUAGCUAUGUUCGGAAAAGAUAACCGCUGAAAGCAUCUAAGCGGGAAACU .(((((((..(.(((.(((((((.(......)))))))).))))(((((((..(((...)))....(((..((....))..)))....))))))))))....((......)))))).... (-40.55 = -40.72 + 0.17)

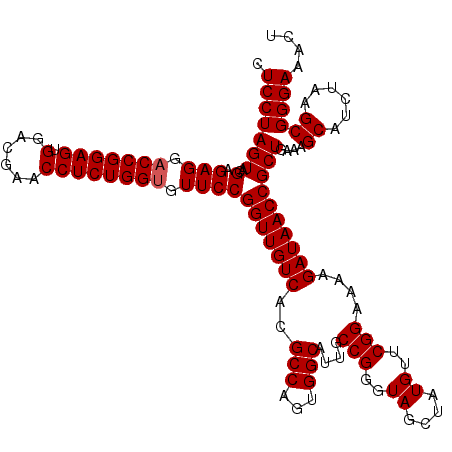
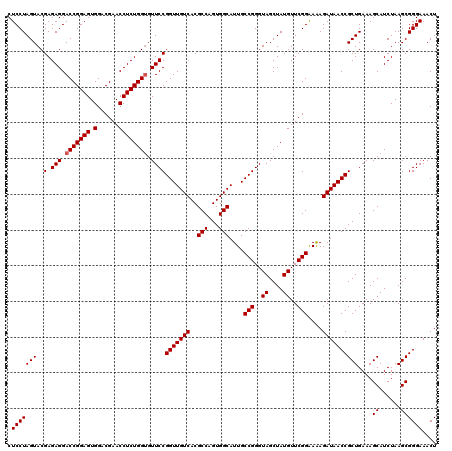
| Location | 4,789,091 – 4,789,210 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.72 |
| Mean single sequence MFE | -32.32 |
| Consensus MFE | -31.92 |
| Energy contribution | -31.92 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.86 |
| SVM RNA-class probability | 0.997417 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4789091 119 + 6264404 CAGCCCCUCUCAAAUCUCAAACGUCCACGG-AGAUAGGGACCGAACUGUCUCACGACGUUCUAAACCCAGCUCGCGUACCACUUUAAAUGGCGAACAGCCAUACCCUUGGGACCGGCUUC .((((..((((((......((((((....(-((((((........)))))))..))))))...........................(((((.....)))))....))))))..)))).. ( -34.90) >pel.2704 4999008 120 + 5888780 CAGCCCCUCUCAAAUCUCAAACGUCCACGGCAGAUAGGGACCGAACUGUCUCACGACGUUCUAAACCCAGCUCGCGUACCACUUUAAAUGGCGAACAGCCAUACCCUUGGGACCGGCUUC .((((..((((((((((....((....))..)))).(((...((((.(((....)))))))..........................(((((.....))))).)))))))))..)))).. ( -31.80) >ppk.338 5479378 120 - 6181863 CAGCCCCUCUCAAAUCUCAAACGUCCACGGCAGAUAGGGACCGAACUGUCUCACGACGUUCUAAACCCAGCUCGCGUACCACUUUAAAUGGCGAACAGCCAUACCCUUGGGACCGGCUUC .((((..((((((((((....((....))..)))).(((...((((.(((....)))))))..........................(((((.....))))).)))))))))..)))).. ( -31.80) >pf5.74 6947389 120 - 7074893 CAGCCCCUCUCAAAUCUCAAACGUCCACGGCAGAUAGGGACCGAACUGUCUCACGACGUUCUAAACCCAGCUCGCGUACCACUUUAAAUGGCGAACAGCCAUACCCUUGGGACCGGCUUC .((((..((((((((((....((....))..)))).(((...((((.(((....)))))))..........................(((((.....))))).)))))))))..)))).. ( -31.80) >pfo.66 6312259 120 - 6438405 CAGCCCCUCUCAAAUCUCAAACGUCCACGGCAGAUAGGGACCGAACUGUCUCACGACGUUCUAAACCCAGCUCGCGUACCACUUUAAAUGGCGAACAGCCAUACCCUUGGGACCGGCUUC .((((..((((((((((....((....))..)))).(((...((((.(((....)))))))..........................(((((.....))))).)))))))))..)))).. ( -31.80) >psp.2973 5779643 120 + 5928787 CAGCCCCUCUCAAAUCUCAAACGUCCACGGCAGAUAGGGACCGAACUGUCUCACGACGUUCUAAACCCAGCUCGCGUACCACUUUAAAUGGCGAACAGCCAUACCCUUGGGACCGGCUUC .((((..((((((((((....((....))..)))).(((...((((.(((....)))))))..........................(((((.....))))).)))))))))..)))).. ( -31.80) >consensus CAGCCCCUCUCAAAUCUCAAACGUCCACGGCAGAUAGGGACCGAACUGUCUCACGACGUUCUAAACCCAGCUCGCGUACCACUUUAAAUGGCGAACAGCCAUACCCUUGGGACCGGCUUC .((((..((((((((((....((....))..)))).(((...((((.(((....)))))))..........................(((((.....))))).)))))))))..)))).. (-31.92 = -31.92 + 0.00)

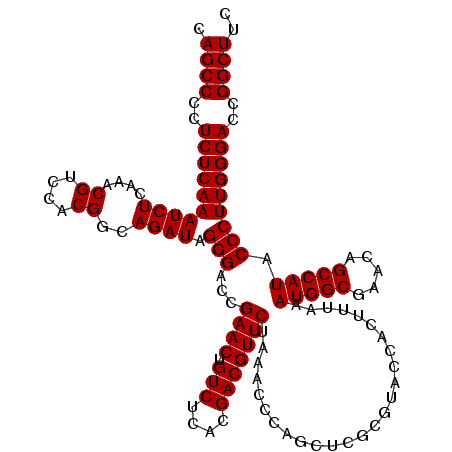
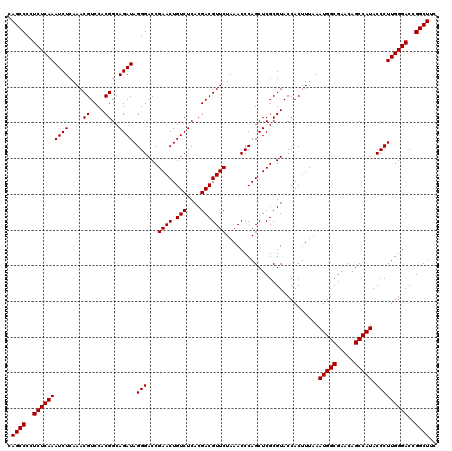
| Location | 4,789,130 – 4,789,250 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -47.30 |
| Consensus MFE | -47.30 |
| Energy contribution | -47.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.64 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.93 |
| SVM RNA-class probability | 0.997804 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4789130 120 - 6264404 UGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUUAAAGUGGUACGCGAGCUGGGUUUAGAACGUCGUGAGACAGUUCGG .........((......((((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....))))))).....(((((((....))).)))))) ( -47.30) >pel.2704 4999048 120 - 5888780 UGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUUAAAGUGGUACGCGAGCUGGGUUUAGAACGUCGUGAGACAGUUCGG .........((......((((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....))))))).....(((((((....))).)))))) ( -47.30) >ppk.338 5479418 120 + 6181863 UGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUUAAAGUGGUACGCGAGCUGGGUUUAGAACGUCGUGAGACAGUUCGG .........((......((((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....))))))).....(((((((....))).)))))) ( -47.30) >pf5.74 6947429 120 + 7074893 UGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUUAAAGUGGUACGCGAGCUGGGUUUAGAACGUCGUGAGACAGUUCGG .........((......((((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....))))))).....(((((((....))).)))))) ( -47.30) >pfo.66 6312299 120 + 6438405 UGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUUAAAGUGGUACGCGAGCUGGGUUUAGAACGUCGUGAGACAGUUCGG .........((......((((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....))))))).....(((((((....))).)))))) ( -47.30) >psp.2973 5779683 120 - 5928787 UGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUUAAAGUGGUACGCGAGCUGGGUUUAGAACGUCGUGAGACAGUUCGG .........((......((((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....))))))).....(((((((....))).)))))) ( -47.30) >consensus UGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUUAAAGUGGUACGCGAGCUGGGUUUAGAACGUCGUGAGACAGUUCGG .........((......((((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....))))))).....(((((((....))).)))))) (-47.30 = -47.30 + 0.00)

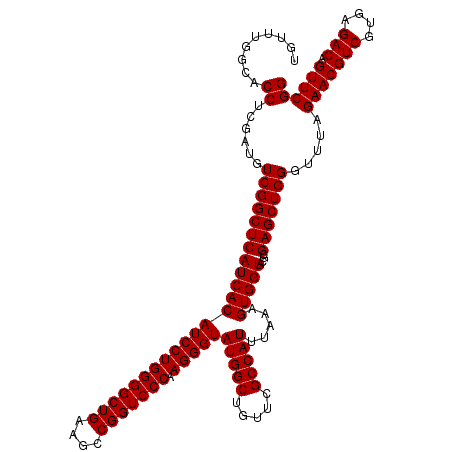
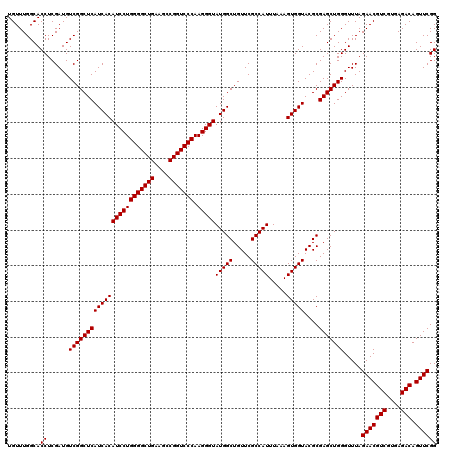
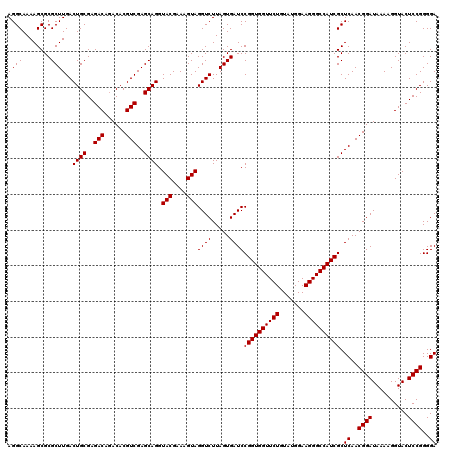
| Location | 4,789,170 – 4,789,290 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -47.20 |
| Consensus MFE | -47.20 |
| Energy contribution | -47.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.27 |
| Structure conservation index | 1.00 |
| SVM decision value | 4.23 |
| SVM RNA-class probability | 0.999842 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4789170 120 - 6264404 UAACAGGCUGAUACCGCCCAAGAGUUCAUAUCGACGGCGGUGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUUAAAGU .....(((.((.((.(((...((((..(((((((....((((.....)))))))))))..)))).....((((((((((((....))))))).)))))..))).)))))))......... ( -47.20) >pel.2704 4999088 120 - 5888780 UAACAGGCUGAUACCGCCCAAGAGUUCAUAUCGACGGCGGUGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUUAAAGU .....(((.((.((.(((...((((..(((((((....((((.....)))))))))))..)))).....((((((((((((....))))))).)))))..))).)))))))......... ( -47.20) >ppk.338 5479458 120 + 6181863 UAACAGGCUGAUACCGCCCAAGAGUUCAUAUCGACGGCGGUGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUUAAAGU .....(((.((.((.(((...((((..(((((((....((((.....)))))))))))..)))).....((((((((((((....))))))).)))))..))).)))))))......... ( -47.20) >pf5.74 6947469 120 + 7074893 UAACAGGCUGAUACCGCCCAAGAGUUCAUAUCGACGGCGGUGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUUAAAGU .....(((.((.((.(((...((((..(((((((....((((.....)))))))))))..)))).....((((((((((((....))))))).)))))..))).)))))))......... ( -47.20) >pfo.66 6312339 120 + 6438405 UAACAGGCUGAUACCGCCCAAGAGUUCAUAUCGACGGCGGUGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUUAAAGU .....(((.((.((.(((...((((..(((((((....((((.....)))))))))))..)))).....((((((((((((....))))))).)))))..))).)))))))......... ( -47.20) >psp.2973 5779723 120 - 5928787 UAACAGGCUGAUACCGCCCAAGAGUUCAUAUCGACGGCGGUGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUUAAAGU .....(((.((.((.(((...((((..(((((((....((((.....)))))))))))..)))).....((((((((((((....))))))).)))))..))).)))))))......... ( -47.20) >consensus UAACAGGCUGAUACCGCCCAAGAGUUCAUAUCGACGGCGGUGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUUAAAGU .....(((.((.((.(((...((((..(((((((....((((.....)))))))))))..)))).....((((((((((((....))))))).)))))..))).)))))))......... (-47.20 = -47.20 + -0.00)
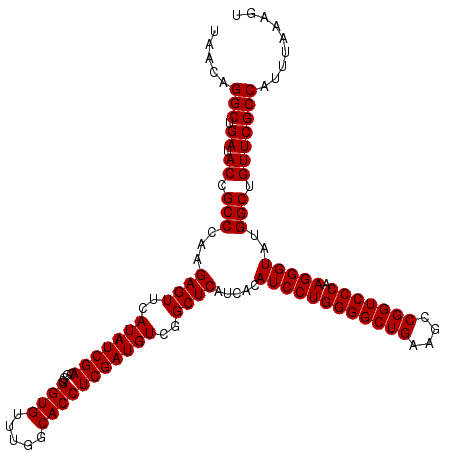
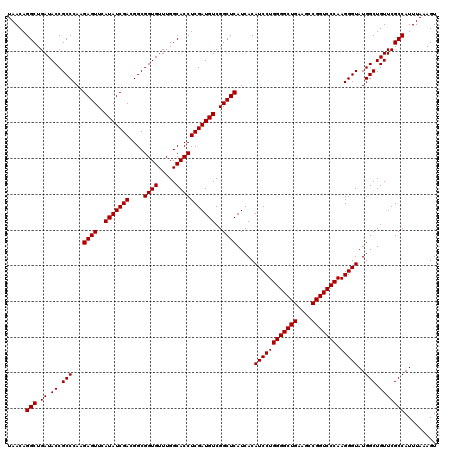
| Location | 4,789,250 – 4,789,370 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -39.10 |
| Consensus MFE | -39.10 |
| Energy contribution | -39.10 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.04 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.940079 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4789250 120 - 6264404 AGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGGUACUCCGGGGAUAACAGGCUGAUACCGCCCAAGAGUUCAUAUCGACGGCGG ...(((....)))(((((..((.(((((((((((((.......)))))))))......((((..........)))).)))).)))))))....(((((...((.......))...))))) ( -39.10) >pel.2704 4999168 120 - 5888780 AGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGGUACUCCGGGGAUAACAGGCUGAUACCGCCCAAGAGUUCAUAUCGACGGCGG ...(((....)))(((((..((.(((((((((((((.......)))))))))......((((..........)))).)))).)))))))....(((((...((.......))...))))) ( -39.10) >ppk.338 5479538 120 + 6181863 AGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGGUACUCCGGGGAUAACAGGCUGAUACCGCCCAAGAGUUCAUAUCGACGGCGG ...(((....)))(((((..((.(((((((((((((.......)))))))))......((((..........)))).)))).)))))))....(((((...((.......))...))))) ( -39.10) >pf5.74 6947549 120 + 7074893 AGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGGUACUCCGGGGAUAACAGGCUGAUACCGCCCAAGAGUUCAUAUCGACGGCGG ...(((....)))(((((..((.(((((((((((((.......)))))))))......((((..........)))).)))).)))))))....(((((...((.......))...))))) ( -39.10) >pfo.66 6312419 120 + 6438405 AGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGGUACUCCGGGGAUAACAGGCUGAUACCGCCCAAGAGUUCAUAUCGACGGCGG ...(((....)))(((((..((.(((((((((((((.......)))))))))......((((..........)))).)))).)))))))....(((((...((.......))...))))) ( -39.10) >psp.2973 5779803 120 - 5928787 AGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGGUACUCCGGGGAUAACAGGCUGAUACCGCCCAAGAGUUCAUAUCGACGGCGG ...(((....)))(((((..((.(((((((((((((.......)))))))))......((((..........)))).)))).)))))))....(((((...((.......))...))))) ( -39.10) >consensus AGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGGUACUCCGGGGAUAACAGGCUGAUACCGCCCAAGAGUUCAUAUCGACGGCGG ...(((....)))(((((..((.(((((((((((((.......)))))))))......((((..........)))).)))).)))))))....(((((...((.......))...))))) (-39.10 = -39.10 + 0.00)

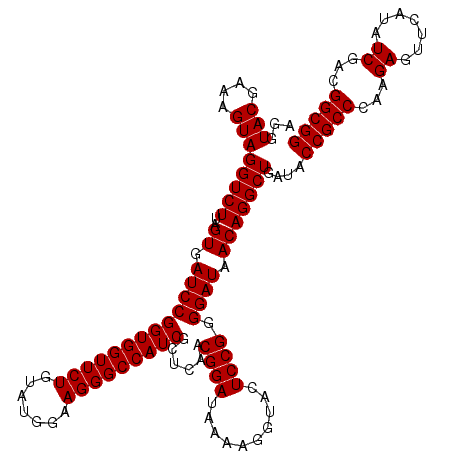
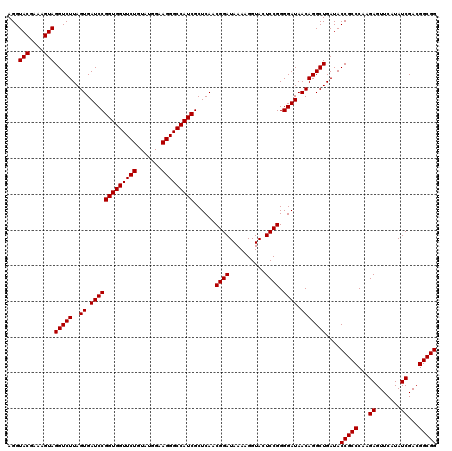
| Location | 4,789,290 – 4,789,410 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.56 |
| Mean single sequence MFE | -38.30 |
| Consensus MFE | -38.30 |
| Energy contribution | -38.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.89 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.877985 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4789290 120 - 6264404 AGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGGUACUCCGGGGA ((((........))))..((((..(((......)))..)))).(((....)))((((.....))))((((((((((.......)))))))))).((..((((..........))))..)) ( -38.30) >pel.2704 4999208 120 - 5888780 AGGCAAAAGCGCGCUUGACUGCGAGACAAACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGGUACUCCGGGGA ((((........))))..((((..(((......)))..)))).(((....)))((((.....))))((((((((((.......)))))))))).((..((((..........))))..)) ( -38.30) >ppk.338 5479578 120 + 6181863 AGGCAAAAGCGCGCUUGACUGCGAGACAAACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGGUACUCCGGGGA ((((........))))..((((..(((......)))..)))).(((....)))((((.....))))((((((((((.......)))))))))).((..((((..........))))..)) ( -38.30) >pf5.74 6947589 120 + 7074893 AGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGGUACUCCGGGGA ((((........))))..((((..(((......)))..)))).(((....)))((((.....))))((((((((((.......)))))))))).((..((((..........))))..)) ( -38.30) >pfo.66 6312459 120 + 6438405 AGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGGUACUCCGGGGA ((((........))))..((((..(((......)))..)))).(((....)))((((.....))))((((((((((.......)))))))))).((..((((..........))))..)) ( -38.30) >psp.2973 5779843 120 - 5928787 AGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGGUACUCCGGGGA ((((........))))..((((..(((......)))..)))).(((....)))((((.....))))((((((((((.......)))))))))).((..((((..........))))..)) ( -38.30) >consensus AGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGGUACUCCGGGGA ((((........))))..((((..(((......)))..)))).(((....)))((((.....))))((((((((((.......)))))))))).((..((((..........))))..)) (-38.30 = -38.30 + 0.00)

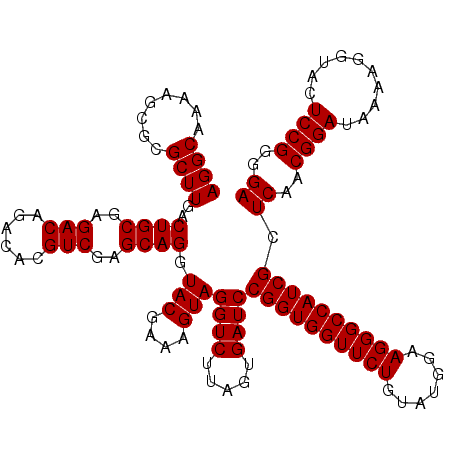
| Location | 4,789,330 – 4,789,450 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.11 |
| Mean single sequence MFE | -39.10 |
| Consensus MFE | -38.90 |
| Energy contribution | -38.90 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.91 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.885843 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4789330 120 - 6264404 GGUGCGCUCAGACCGGUCGGAAAUCGGUCGCAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAU ((((((((..(((((((.....)))))))((...........))...))))))))...((((..(((......)))..)))).(((....)))((((.....))))(((.....)))... ( -39.50) >pau.414 5800161 120 + 6537648 GGUGCGCUCAGACCGGUCGGAAAUCGGUCGCAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAU ((((((((..(((((((.....)))))))((...........))...))))))))...((((..(((......)))..)))).(((....)))((((.....))))(((.....)))... ( -39.50) >pel.2704 4999248 120 - 5888780 GGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAAACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAU ((((((((..(((((((.....))))))).....((......))...))))))))...((((..(((......)))..)))).(((....)))((((.....))))(((.....)))... ( -38.90) >ppk.338 5479618 120 + 6181863 GGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAAACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAU ((((((((..(((((((.....))))))).....((......))...))))))))...((((..(((......)))..)))).(((....)))((((.....))))(((.....)))... ( -38.90) >pfo.66 6312499 120 + 6438405 GGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAU ((((((((..(((((((.....))))))).....((......))...))))))))...((((..(((......)))..)))).(((....)))((((.....))))(((.....)))... ( -38.90) >psp.2973 5779883 120 - 5928787 GGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAU ((((((((..(((((((.....))))))).....((......))...))))))))...((((..(((......)))..)))).(((....)))((((.....))))(((.....)))... ( -38.90) >consensus GGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAU ((((((((..(((((((.....))))))).....((......))...))))))))...((((..(((......)))..)))).(((....)))((((.....))))(((.....)))... (-38.90 = -38.90 + -0.00)

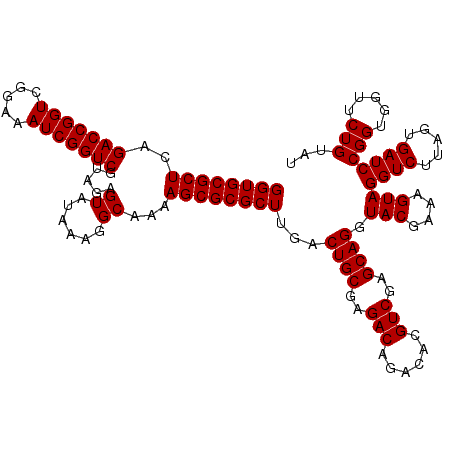
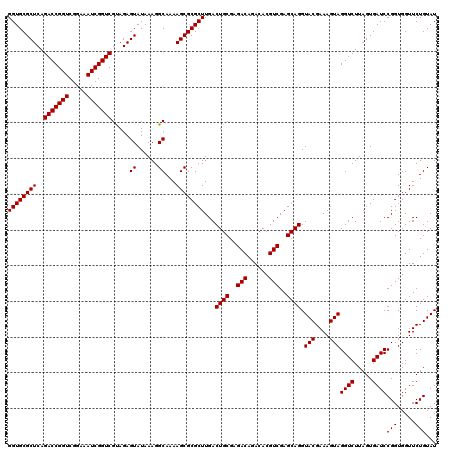
| Location | 4,789,370 – 4,789,490 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.67 |
| Mean single sequence MFE | -36.89 |
| Consensus MFE | -35.77 |
| Energy contribution | -35.77 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.976800 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4789370 120 + 6264404 GCUCGACGUGUCUGUCUCGCAGUCAAGCGCGCUUUUGCCUUUAUACUCUGCGACCGAUUUCCGACCGGUCUGAGCGCACCUUCGUACUCCUCCGUUACUCUUUAGGAGGAGACCGCCCCA (((.((((((.......))).))).)))((((((..((...........))(((((.........))))).)))))).....((..(((((((...........)))))))..))..... ( -38.10) >pau.414 5800201 120 - 6537648 GCUCGACGUGUCUGUCUCGCAGUCAAGCGCGCUUUUGCCUUUAUACUCUGCGACCGAUUUCCGACCGGUCUGAGCGCACCUUCGUACUCCUCCGUUACUCUUUAGGAGGAGACCGCCCCA (((.((((((.......))).))).)))((((((..((...........))(((((.........))))).)))))).....((..(((((((...........)))))))..))..... ( -38.10) >pel.2704 4999288 120 + 5888780 GCUCGACGUGUUUGUCUCGCAGUCAAGCGCGCUUUUGCCUUUAUACUCUACGACCGAUUUCCGACCGGUCUGAGCGCACCUUCGUACUCCUCCGUUACUCUUUGGGAGGAGACCGCCCCA (((((.((((((((.(.....).)))))))))...................(((((.........))))).)))).......((..(((((((...........)))))))..))..... ( -36.60) >ppk.338 5479658 120 - 6181863 GCUCGACGUGUUUGUCUCGCAGUCAAGCGCGCUUUUGCCUUUAUACUCUACGACCGAUUUCCGACCGGUCUGAGCGCACCUUCGUACUCCUCCGUUACUCUUUGGGAGGAGACCGCCCCA (((((.((((((((.(.....).)))))))))...................(((((.........))))).)))).......((..(((((((...........)))))))..))..... ( -36.60) >pfo.66 6312539 120 - 6438405 GCUCGACGUGUCUGUCUCGCAGUCAAGCGCGCUUUUGCCUUUAUACUCUACGACCGAUUUCCGACCGGUCUGAGCGCACCUUCGUACUCCUCCGUUACUCUUUAGGAGGAGACCGCCCCA (((.((((((.......))).))).)))((((((.................(((((.........))))).)))))).....((..(((((((...........)))))))..))..... ( -35.97) >psp.2973 5779923 120 + 5928787 GCUCGACGUGUCUGUCUCGCAGUCAAGCGCGCUUUUGCCUUUAUACUCUACGACCGAUUUCCGACCGGUCUGAGCGCACCUUCGUACUCCUCCGUUACUCUUUAGGAGGAGACCGCCCCA (((.((((((.......))).))).)))((((((.................(((((.........))))).)))))).....((..(((((((...........)))))))..))..... ( -35.97) >consensus GCUCGACGUGUCUGUCUCGCAGUCAAGCGCGCUUUUGCCUUUAUACUCUACGACCGAUUUCCGACCGGUCUGAGCGCACCUUCGUACUCCUCCGUUACUCUUUAGGAGGAGACCGCCCCA (((.((((((.......))).))).)))((((((.................(((((.........))))).)))))).....((..(((((((...........)))))))..))..... (-35.77 = -35.77 + -0.00)
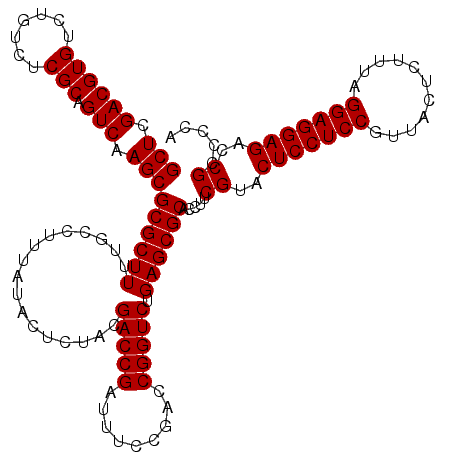
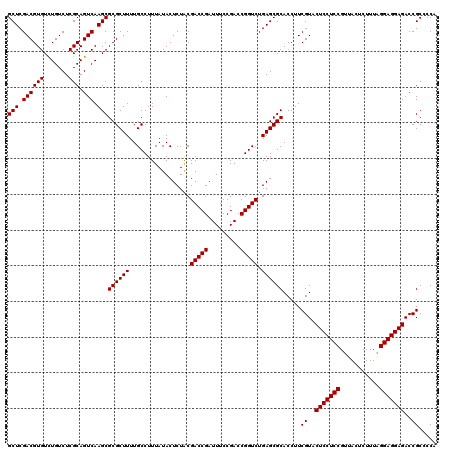
| Location | 4,789,370 – 4,789,490 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.67 |
| Mean single sequence MFE | -44.27 |
| Consensus MFE | -43.93 |
| Energy contribution | -43.93 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.99 |
| SVM decision value | 3.81 |
| SVM RNA-class probability | 0.999634 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4789370 120 - 6264404 UGGGGCGGUCUCCUCCUAAAGAGUAACGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGCAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGAGC ....((((((((((((...........))))))......(((((((((..(((((((.....)))))))((...........))...)))))))))))))))..(((......))).... ( -44.70) >pau.414 5800201 120 + 6537648 UGGGGCGGUCUCCUCCUAAAGAGUAACGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGCAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGAGC ....((((((((((((...........))))))......(((((((((..(((((((.....)))))))((...........))...)))))))))))))))..(((......))).... ( -44.70) >pel.2704 4999288 120 - 5888780 UGGGGCGGUCUCCUCCCAAAGAGUAACGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAAACACGUCGAGC ....((((((((((((...........))))))......(((((((((..(((((((.....))))))).....((......))...)))))))))))))))..(((......))).... ( -43.60) >ppk.338 5479658 120 + 6181863 UGGGGCGGUCUCCUCCCAAAGAGUAACGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAAACACGUCGAGC ....((((((((((((...........))))))......(((((((((..(((((((.....))))))).....((......))...)))))))))))))))..(((......))).... ( -43.60) >pfo.66 6312539 120 + 6438405 UGGGGCGGUCUCCUCCUAAAGAGUAACGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGAGC ..(.((.(((((((((...........))))))((((....))))((((.(((((((.....)))))))...)))).....)))....)).)((((((((((......).)).))))))) ( -44.50) >psp.2973 5779923 120 - 5928787 UGGGGCGGUCUCCUCCUAAAGAGUAACGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGAGC ..(.((.(((((((((...........))))))((((....))))((((.(((((((.....)))))))...)))).....)))....)).)((((((((((......).)).))))))) ( -44.50) >consensus UGGGGCGGUCUCCUCCUAAAGAGUAACGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGAGC ....((((((((((((...........))))))......(((((((((..(((((((.....))))))).....((......))...)))))))))))))))..(((......))).... (-43.93 = -43.93 + -0.00)

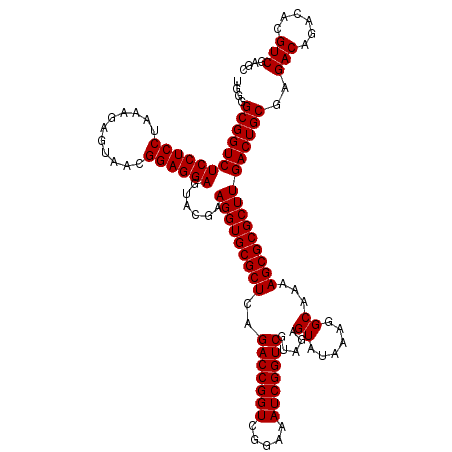
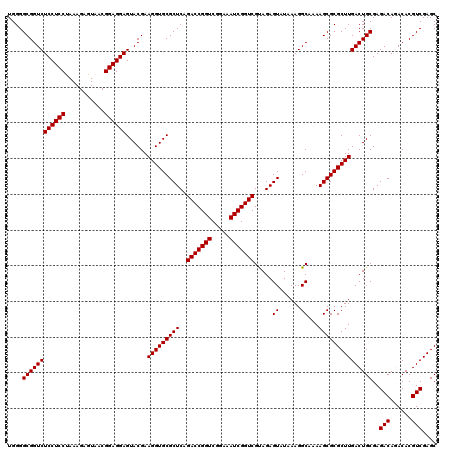
| Location | 4,789,410 – 4,789,530 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.17 |
| Mean single sequence MFE | -27.47 |
| Consensus MFE | -26.50 |
| Energy contribution | -26.50 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.01 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.874174 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4789410 120 + 6264404 UUAUACUCUGCGACCGAUUUCCGACCGGUCUGAGCGCACCUUCGUACUCCUCCGUUACUCUUUAGGAGGAGACCGCCCCAGUCAAACUGCCCACCAUACACUGUCCUCGAUCCGGAUUAC ........................((((..((((.(((....((..(((((((...........)))))))..))...(((.....)))............))).))))..))))..... ( -29.00) >pau.414 5800241 120 - 6537648 UUAUACUCUGCGACCGAUUUCCGACCGGUCUGAGCGCACCUUCGUACUCCUCCGUUACUCUUUAGGAGGAGACCGCCCCAGUCAAACUGCCCACCAUACACUGUCCUCGAUCCGGAUCAC ......................((((((..((((.(((....((..(((((((...........)))))))..))...(((.....)))............))).))))..)))).)).. ( -30.20) >pel.2704 4999328 120 + 5888780 UUAUACUCUACGACCGAUUUCCGACCGGUCUGAGCGCACCUUCGUACUCCUCCGUUACUCUUUGGGAGGAGACCGCCCCAGUCAAACUACCCACCAUACACUGUCCUCGAUCCGGAUAAC ........................((((..((((.(((....((..(((((((...........)))))))..))....((.....)).............))).))))..))))..... ( -26.10) >ppk.338 5479698 120 - 6181863 UUAUACUCUACGACCGAUUUCCGACCGGUCUGAGCGCACCUUCGUACUCCUCCGUUACUCUUUGGGAGGAGACCGCCCCAGUCAAACUACCCACCAUACACUGUCCUCGAUCCGGAUAAC ........................((((..((((.(((....((..(((((((...........)))))))..))....((.....)).............))).))))..))))..... ( -26.10) >pfo.66 6312579 120 - 6438405 UUAUACUCUACGACCGAUUUCCGACCGGUCUGAGCGCACCUUCGUACUCCUCCGUUACUCUUUAGGAGGAGACCGCCCCAGUCAAACUACCCACCAUACACUGUCCUCGAUCCGGAUAAC ........................((((..((((.(((....((..(((((((...........)))))))..))....((.....)).............))).))))..))))..... ( -26.70) >psp.2973 5779963 120 + 5928787 UUAUACUCUACGACCGAUUUCCGACCGGUCUGAGCGCACCUUCGUACUCCUCCGUUACUCUUUAGGAGGAGACCGCCCCAGUCAAACUACCCACCAUACACUGUCCUCGAUCCGGAUAAC ........................((((..((((.(((....((..(((((((...........)))))))..))....((.....)).............))).))))..))))..... ( -26.70) >consensus UUAUACUCUACGACCGAUUUCCGACCGGUCUGAGCGCACCUUCGUACUCCUCCGUUACUCUUUAGGAGGAGACCGCCCCAGUCAAACUACCCACCAUACACUGUCCUCGAUCCGGAUAAC ........................((((..((((.(((....((..(((((((...........)))))))..))....((.....)).............))).))))..))))..... (-26.50 = -26.50 + -0.00)

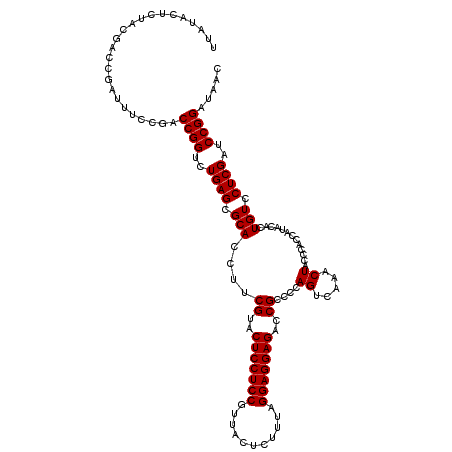
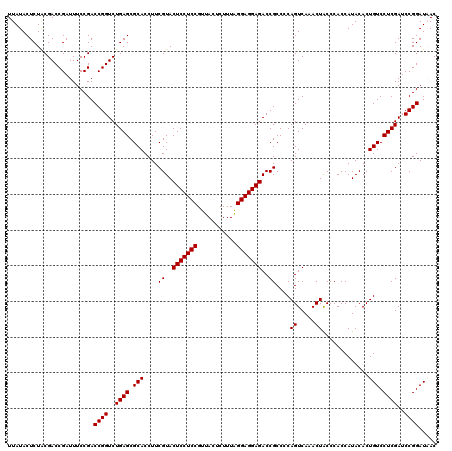
| Location | 4,789,450 – 4,789,570 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.78 |
| Mean single sequence MFE | -36.27 |
| Consensus MFE | -33.73 |
| Energy contribution | -33.73 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.56 |
| SVM RNA-class probability | 0.995255 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4789450 120 + 6264404 UUCGUACUCCUCCGUUACUCUUUAGGAGGAGACCGCCCCAGUCAAACUGCCCACCAUACACUGUCCUCGAUCCGGAUUACGGACCAGAGUUAGAACCUCAAGCAUGCCAGGGUGGUAUUU ......(((((((...........)))))))((((((((((.....)))...........(((..(((..((((.....))))...))).)))................))))))).... ( -36.70) >pau.414 5800281 120 - 6537648 UUCGUACUCCUCCGUUACUCUUUAGGAGGAGACCGCCCCAGUCAAACUGCCCACCAUACACUGUCCUCGAUCCGGAUCACGGACCAGAGUUAGAACCUCAAGCAUGCCAGGGUGGUAUUU ......(((((((...........)))))))((((((((((.....)))...........(((..(((..((((.....))))...))).)))................))))))).... ( -36.70) >pel.2704 4999368 120 + 5888780 UUCGUACUCCUCCGUUACUCUUUGGGAGGAGACCGCCCCAGUCAAACUACCCACCAUACACUGUCCUCGAUCCGGAUAACGGACCUGAGUUAGAACCUCAAGGUUGCCAGGGUGGUAUUU ......(((((((...........)))))))(((((((..((..................(((..((((.((((.....))))..)))).)))((((....))))))..))))))).... ( -38.30) >ppk.338 5479738 120 - 6181863 UUCGUACUCCUCCGUUACUCUUUGGGAGGAGACCGCCCCAGUCAAACUACCCACCAUACACUGUCCUCGAUCCGGAUAACGGACCUGAGUUAGAACCUCAAAGUUGCCAGGGUGGUAUUU ......(((((((...........)))))))(((((((..((..((((....................(.((((.....)))).)((((.......)))).))))))..))))))).... ( -34.90) >pfo.66 6312619 120 - 6438405 UUCGUACUCCUCCGUUACUCUUUAGGAGGAGACCGCCCCAGUCAAACUACCCACCAUACACUGUCCUCGAUCCGGAUAACGGACCUGAGUUAGAACCUCAAAGUUGCCAGGGUGGUAUUU ......(((((((...........)))))))(((((((..((..((((....................(.((((.....)))).)((((.......)))).))))))..))))))).... ( -35.50) >psp.2973 5780003 120 + 5928787 UUCGUACUCCUCCGUUACUCUUUAGGAGGAGACCGCCCCAGUCAAACUACCCACCAUACACUGUCCUCGAUCCGGAUAACGGACCUGAGUUAGAACCUCAAAGUUGCCAGGGUGGUAUUU ......(((((((...........)))))))(((((((..((..((((....................(.((((.....)))).)((((.......)))).))))))..))))))).... ( -35.50) >consensus UUCGUACUCCUCCGUUACUCUUUAGGAGGAGACCGCCCCAGUCAAACUACCCACCAUACACUGUCCUCGAUCCGGAUAACGGACCUGAGUUAGAACCUCAAAGUUGCCAGGGUGGUAUUU ......(((((((...........)))))))(((((((((((.................))))..((((.((((.....)))).).)))....................))))))).... (-33.73 = -33.73 + 0.00)

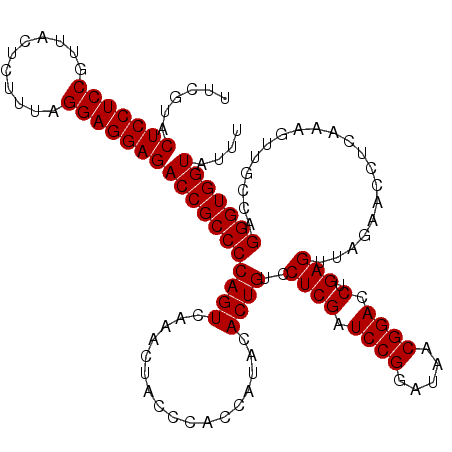
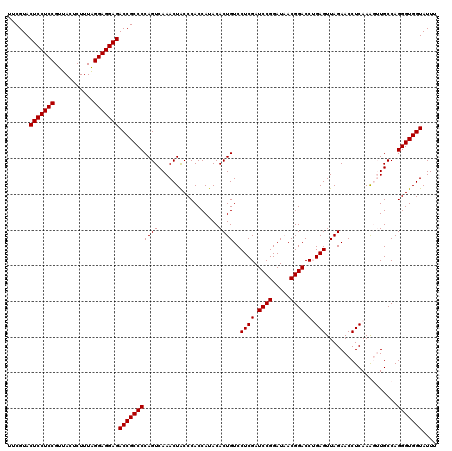
| Location | 4,789,450 – 4,789,570 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.78 |
| Mean single sequence MFE | -39.17 |
| Consensus MFE | -37.49 |
| Energy contribution | -37.35 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.06 |
| Mean z-score | -0.82 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.638659 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4789450 120 - 6264404 AAAUACCACCCUGGCAUGCUUGAGGUUCUAACUCUGGUCCGUAAUCCGGAUCGAGGACAGUGUAUGGUGGGCAGUUUGACUGGGGCGGUCUCCUCCUAAAGAGUAACGGAGGAGUACGAA ....(((.(((..(((((((((..((.((..(((.((((((.....)))))))))...))...))..))))))...)).)..))).)))(((((((...........)))))))...... ( -41.10) >pau.414 5800281 120 + 6537648 AAAUACCACCCUGGCAUGCUUGAGGUUCUAACUCUGGUCCGUGAUCCGGAUCGAGGACAGUGUAUGGUGGGCAGUUUGACUGGGGCGGUCUCCUCCUAAAGAGUAACGGAGGAGUACGAA ....(((.(((..(((((((((..((.((..(((.((((((.....)))))))))...))...))..))))))...)).)..))).)))(((((((...........)))))))...... ( -41.10) >pel.2704 4999368 120 - 5888780 AAAUACCACCCUGGCAACCUUGAGGUUCUAACUCAGGUCCGUUAUCCGGAUCGAGGACAGUGUAUGGUGGGUAGUUUGACUGGGGCGGUCUCCUCCCAAAGAGUAACGGAGGAGUACGAA ....(((.(((..(((((((((((.......))).((((((.....)))))))))).((.....)).........))).)..))).)))(((((((...........)))))))...... ( -39.90) >ppk.338 5479738 120 + 6181863 AAAUACCACCCUGGCAACUUUGAGGUUCUAACUCAGGUCCGUUAUCCGGAUCGAGGACAGUGUAUGGUGGGUAGUUUGACUGGGGCGGUCUCCUCCCAAAGAGUAACGGAGGAGUACGAA ....(((.(((..((((((.((((.......))))((((((.....))))))....................)).))).)..))).)))(((((((...........)))))))...... ( -37.30) >pfo.66 6312619 120 + 6438405 AAAUACCACCCUGGCAACUUUGAGGUUCUAACUCAGGUCCGUUAUCCGGAUCGAGGACAGUGUAUGGUGGGUAGUUUGACUGGGGCGGUCUCCUCCUAAAGAGUAACGGAGGAGUACGAA ....(((.(((..((((((.((((.......))))((((((.....))))))....................)).))).)..))).)))(((((((...........)))))))...... ( -37.80) >psp.2973 5780003 120 - 5928787 AAAUACCACCCUGGCAACUUUGAGGUUCUAACUCAGGUCCGUUAUCCGGAUCGAGGACAGUGUAUGGUGGGUAGUUUGACUGGGGCGGUCUCCUCCUAAAGAGUAACGGAGGAGUACGAA ....(((.(((..((((((.((((.......))))((((((.....))))))....................)).))).)..))).)))(((((((...........)))))))...... ( -37.80) >consensus AAAUACCACCCUGGCAACCUUGAGGUUCUAACUCAGGUCCGUUAUCCGGAUCGAGGACAGUGUAUGGUGGGUAGUUUGACUGGGGCGGUCUCCUCCUAAAGAGUAACGGAGGAGUACGAA .....(((((...(((.(((((((.......))).((((((.....))))))))))....)))..))))).(((.....)))...((..(((((((...........)))))))..)).. (-37.49 = -37.35 + -0.14)

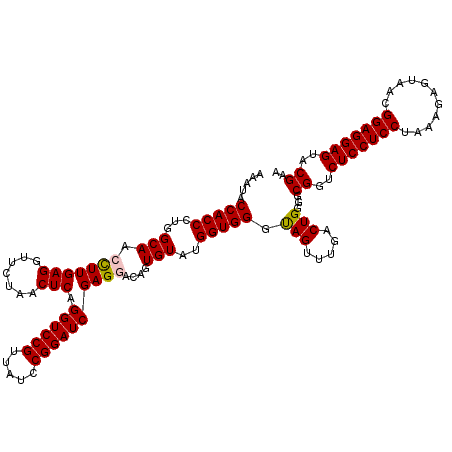
| Location | 4,789,530 – 4,789,650 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.61 |
| Mean single sequence MFE | -42.17 |
| Consensus MFE | -41.20 |
| Energy contribution | -40.03 |
| Covariance contribution | -1.16 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958665 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2545 4789530 120 - 6264404 CACUGGACUUUGAGCCUGCUUGUGUAGGAUAGGUGGGAGGCUUUGAAGCGUGGACGCCAGUUCGCGUGGAGCCAUCCUUGAAAUACCACCCUGGCAUGCUUGAGGUUCUAACUCUGGUCC ....((((((..(((.((((.(.((.((.((...(((((((((....((((((((....)))))))).))))).)))).....)))))).).)))).)))..))))))............ ( -45.80) >pau.414 5800361 120 + 6537648 CACUGGACUUUGAGCCUGCUUGUGUAGGAUAGGUGGGAGGCUUUGAAGCGUGGACGCCAGUUCGCGUGGAGCCAUCCUUGAAAUACCACCCUGGCAUGCUUGAGGUUCUAACUCUGGUCC ....((((((..(((.((((.(.((.((.((...(((((((((....((((((((....)))))))).))))).)))).....)))))).).)))).)))..))))))............ ( -45.80) >pss.3053 5948917 120 - 6093698 CACUGGACUUUGAAUUUGCUUGUGUAGGAUAGGUGGGAGGCUUUGAAGCGUGGACGCCAGUCUGCGUGGAGCCAACCUUGAAAUACCACCCUGGCAACUUUGAGGUUCUAACUCAGGUCC ....((((((..((.(((((.(((((....((((....(((((....(((..(((....)))..))).))))).)))).....)).)))...))))).))..))))))............ ( -40.00) >pel.2704 4999448 120 - 5888780 CACUGGACUUUGAGCUUGCUUGUGUAGGAUAGGUGGGAGGCUUUGAAGUGGGGACGCCAGUUCUCAUGGAGCCAUCCUUGAAAUACCACCCUGGCAACCUUGAGGUUCUAACUCAGGUCC ....((((((..((.(((((.(.((.((.((...(((((((((....((((((((....)))))))).))))).)))).....)))))).).))))).))..))))))............ ( -41.60) >ppk.338 5479818 120 + 6181863 CACUGGACUUUGAAUUUGCUUGUGUAGGAUAGGUGGGAGGCUUUGAAGUGGGGACGCCAGUUCUCAUGGAGCCAUCCUUGAAAUACCACCCUGGCAACUUUGAGGUUCUAACUCAGGUCC ....((((((..((.(((((.(.((.((.((...(((((((((....((((((((....)))))))).))))).)))).....)))))).).))))).))..))))))............ ( -39.80) >psp.2973 5780083 120 - 5928787 CACUGGACUUUGAAUUUGCUUGUGUAGGAUAGGUGGGAGGCUUUGAAGCGUGGACGCCAGUCUGCGUGGAGCCAACCUUGAAAUACCACCCUGGCAACUUUGAGGUUCUAACUCAGGUCC ....((((((..((.(((((.(((((....((((....(((((....(((..(((....)))..))).))))).)))).....)).)))...))))).))..))))))............ ( -40.00) >consensus CACUGGACUUUGAACUUGCUUGUGUAGGAUAGGUGGGAGGCUUUGAAGCGUGGACGCCAGUUCGCGUGGAGCCAUCCUUGAAAUACCACCCUGGCAACCUUGAGGUUCUAACUCAGGUCC ....((((((..((.(((((.(.((.((.((...(((((((((....((((((((....)))))))).))))).)))).....)))))).).))))).))..))))))............ (-41.20 = -40.03 + -1.16)
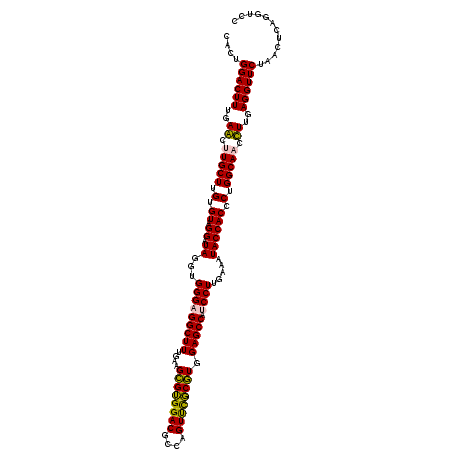
Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:08:51 2007