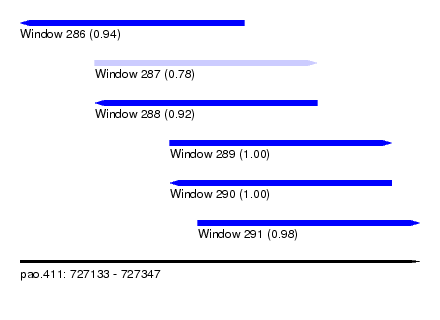
| Sequence ID | pao.411 |
|---|---|
| Location | 727,133 – 727,347 |
| Length | 214 |
| Max. P | 0.999748 |

| Location | 727,133 – 727,253 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.42 |
| Mean single sequence MFE | -40.77 |
| Consensus MFE | -40.35 |
| Energy contribution | -39.60 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937004 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 727133 120 - 6264404 GCUUGACGAUGACCUACUCUCACAUGGGGAGACCCCACACUACCAUCGGCGAUGCGUCGUUUCACUUCUGAGUUCGGGAAGGGAUCAGGUGGUUCCAACGCUCUAUGAUCGUCAAGCAAU ((((((((((.((((...((((...(((....)))...........(((((...))))).........))))...)))..((((((....)))))).........).))))))))))... ( -42.00) >pau.2632 1585501 120 + 6537648 GCUUGACGAUGACCUACUCUCACAUGGGGAGACCCCACACUACCAUCGGCGAUGCGUCGUUUCACUUCUGAGUUCGGGAAGGGAUCAGGUGGUUCCAACGCUCUAUGAUCGUCAAGCAAU ((((((((((.((((...((((...(((....)))...........(((((...))))).........))))...)))..((((((....)))))).........).))))))))))... ( -42.00) >pss.496 969820 120 - 6093698 UCUUGACGAUGACCUACUCUCACAUGGGGAAACCCCACACUACCAUCGGCGAUACAUCGUUUCACUACUGAGUUCGGGAUGGGAUCAGGUGGUUCCAAUGCUCUAUGGUCGUCAAGAAAU ((((((((((.((((...((((...(((....)))............(((((....))))).......))))...))).(((((((....)))))))........).))))))))))... ( -39.30) >pfo.1282 2554627 120 - 6438405 UCUUGACGAUGACCUACUCUCACAUGGGGAAACCCCACACUACCAUCGGCGAUGCAUCGUUUCACUUCUGAGUUCGGGAUGGGAUCAGGUGGUUCCAACGCUCUAUGGUCGUCAAGAAAU (((((((((................(((....))).......((((.((((..............(((((....)))))(((((((....))))))).))).).)))))))))))))... ( -39.80) >consensus GCUUGACGAUGACCUACUCUCACAUGGGGAAACCCCACACUACCAUCGGCGAUGCAUCGUUUCACUUCUGAGUUCGGGAAGGGAUCAGGUGGUUCCAACGCUCUAUGAUCGUCAAGAAAU ((((((((((.((((...((((...(((....)))............(((((....))))).......))))...)))..((((((....)))))).........).))))))))))... (-40.35 = -39.60 + -0.75)


| Location | 727,173 – 727,292 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.42 |
| Mean single sequence MFE | -43.08 |
| Consensus MFE | -28.12 |
| Energy contribution | -28.38 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.777078 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 727173 119 + 6264404 UUCCCGAACUCAGAAGUGAAACGACGCAUCGCCGAUGGUAGUGUGGGGUCUCCCCAUGUGAGAGUAGGUCAUCGUCAAGCUCCUAUCCCAAAACCCCUGGUCAGCGAUG-ACCGGGGGUU ...........((.(((.....((((.((.(((.((....(..(((((...)))))..)....)).))).))))))..))).)).......((((((((((((....))-)))))))))) ( -42.60) >pau.2632 1585541 119 - 6537648 UUCCCGAACUCAGAAGUGAAACGACGCAUCGCCGAUGGUAGUGUGGGGUCUCCCCAUGUGAGAGUAGGUCAUCGUCAAGCUCCUAUCCCAAAACCCCUGGUCAGCGAUG-ACCAGGGGUU ...........((.(((.....((((.((.(((.((....(..(((((...)))))..)....)).))).))))))..))).)).......((((((((((((....))-)))))))))) ( -43.30) >pss.496 969860 120 + 6093698 AUCCCGAACUCAGUAGUGAAACGAUGUAUCGCCGAUGGUAGUGUGGGGUUUCCCCAUGUGAGAGUAGGUCAUCGUCAAGAUUGAAUUCCAGAAACCCUCAUCGCUUACGCGUUGAGGGUU .((..(((.(((((..(((..(((((....(((.((....(..(((((...)))))..)....)).)))))))))))..))))).)))..))((((((((.(((....))).)))))))) ( -44.00) >pfo.1282 2554667 119 + 6438405 AUCCCGAACUCAGAAGUGAAACGAUGCAUCGCCGAUGGUAGUGUGGGGUUUCCCCAUGUGAGAGUAGGUCAUCGUCAAGAUUAAAUUCC-GAAACCCCAAUUGCGAAAGCAGUUGGGGUU .........((.(((...((((((((....(((.((....(..(((((...)))))..)....)).))))))))).....))...))).-))((((((((((((....)))))))))))) ( -42.40) >consensus AUCCCGAACUCAGAAGUGAAACGACGCAUCGCCGAUGGUAGUGUGGGGUCUCCCCAUGUGAGAGUAGGUCAUCGUCAAGAUCCAAUCCCAAAAACCCUGAUCACCGAUG_ACCGGGGGUU ......................((((.((.(((.((....(..(((((...)))))..)....)).))).))))))...............((((((((((((....)).)))))))))) (-28.12 = -28.38 + 0.25)


| Location | 727,173 – 727,292 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.42 |
| Mean single sequence MFE | -42.30 |
| Consensus MFE | -27.94 |
| Energy contribution | -29.25 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.917371 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 727173 119 - 6264404 AACCCCCGGU-CAUCGCUGACCAGGGGUUUUGGGAUAGGAGCUUGACGAUGACCUACUCUCACAUGGGGAGACCCCACACUACCAUCGGCGAUGCGUCGUUUCACUUCUGAGUUCGGGAA .(((((.(((-((....))))).)))))(((.(((((((((...(((((((..............(((....)))........((((...)))))))))))...)))))..)))).))). ( -40.60) >pau.2632 1585541 119 + 6537648 AACCCCUGGU-CAUCGCUGACCAGGGGUUUUGGGAUAGGAGCUUGACGAUGACCUACUCUCACAUGGGGAGACCCCACACUACCAUCGGCGAUGCGUCGUUUCACUUCUGAGUUCGGGAA .(((((((((-((....)))))))))))(((.(((((((((...(((((((..............(((....)))........((((...)))))))))))...)))))..)))).))). ( -45.20) >pss.496 969860 120 - 6093698 AACCCUCAACGCGUAAGCGAUGAGGGUUUCUGGAAUUCAAUCUUGACGAUGACCUACUCUCACAUGGGGAAACCCCACACUACCAUCGGCGAUACAUCGUUUCACUACUGAGUUCGGGAU ((((((((.(((....))).))))))))(((.(((((((.....(((((((..............(((....)))..(.((......)).)...))))))).......))))))).))). ( -45.10) >pfo.1282 2554667 119 - 6438405 AACCCCAACUGCUUUCGCAAUUGGGGUUUC-GGAAUUUAAUCUUGACGAUGACCUACUCUCACAUGGGGAAACCCCACACUACCAUCGGCGAUGCAUCGUUUCACUUCUGAGUUCGGGAU .(((((((.(((....))).)))))))(((-.(((((((.....(((((((..............(((....)))........((((...))))))))))).......))))))).))). ( -38.30) >consensus AACCCCCAGU_CAUCGCCGACCAGGGGUUCUGGAAUAGAAGCUUGACGAUGACCUACUCUCACAUGGGGAAACCCCACACUACCAUCGGCGAUGCAUCGUUUCACUUCUGAGUUCGGGAA ..((((((.(((....))).))))))..(((.(((((((.....(((((((..............(((....)))........((((...))))))))))).......))))))).))). (-27.94 = -29.25 + 1.31)

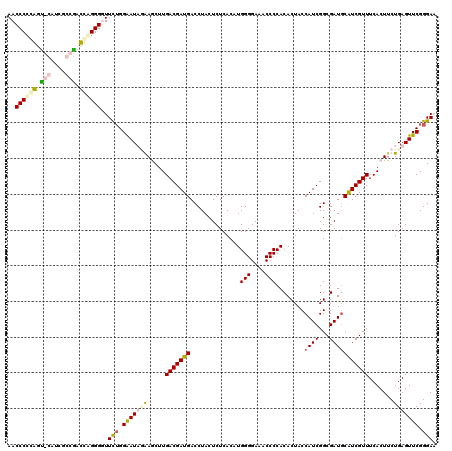
| Location | 727,213 – 727,332 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 64.67 |
| Mean single sequence MFE | -44.15 |
| Consensus MFE | -22.52 |
| Energy contribution | -24.02 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.22 |
| Mean z-score | -4.01 |
| Structure conservation index | 0.51 |
| SVM decision value | 4.00 |
| SVM RNA-class probability | 0.999748 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 727213 119 + 6264404 GUGUGGGGUCUCCCCAUGUGAGAGUAGGUCAUCGUCAAGCUCCUAUCCCAAAACCCCUGGUCAGCGAUG-ACCGGGGGUUUUGCUUUUGCGCGCGGAAAAAGGCGUGCUUCCUGUUCACC ..((((((...))))))(((((..((((....................(((((((((((((((....))-))))))))))))).....((((((........))))))..))))))))). ( -51.80) >pau.2632 1585581 119 - 6537648 GUGUGGGGUCUCCCCAUGUGAGAGUAGGUCAUCGUCAAGCUCCUAUCCCAAAACCCCUGGUCAGCGAUG-ACCAGGGGUUUUGCUUUUGCGCGCGGAAAAAGGCGUGCUUCCUGUUCACC ..((((((...))))))(((((..((((....................(((((((((((((((....))-))))))))))))).....((((((........))))))..))))))))). ( -52.50) >pss.496 969900 86 + 6093698 GUGUGGGGUUUCCCCAUGUGAGAGUAGGUCAUCGUCAAGAUUGAAUUCCAGAAACCCUCAUCGCUUACGCGUUGAGGGUUUUUGUU---------------------------------- (..(((((...)))))..)..((((.((((........))))..))))((((((((((((.(((....))).))))))))))))..---------------------------------- ( -34.40) >pfo.1282 2554707 85 + 6438405 GUGUGGGGUUUCCCCAUGUGAGAGUAGGUCAUCGUCAAGAUUAAAUUCC-GAAACCCCAAUUGCGAAAGCAGUUGGGGUUUUGUUU---------------------------------- (..(((((...)))))..)..((((.((((........))))..))))(-((((((((((((((....)))))))))))))))...---------------------------------- ( -37.90) >consensus GUGUGGGGUCUCCCCAUGUGAGAGUAGGUCAUCGUCAAGAUCCAAUCCCAAAAACCCUGAUCACCGAUG_ACCGGGGGUUUUGCUU__________________________________ ..((((((...)))))).(((((........)).)))...........(((((((((((((((....)).))))))))))))).....((((((........))))))............ (-22.52 = -24.02 + 1.50)

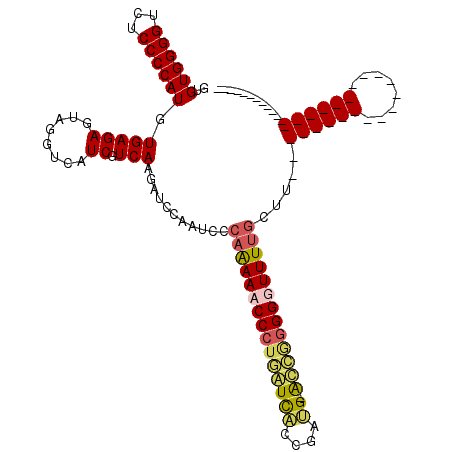
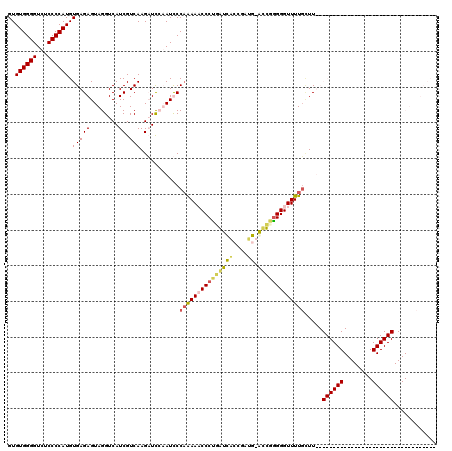
| Location | 727,213 – 727,332 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 64.67 |
| Mean single sequence MFE | -36.85 |
| Consensus MFE | -15.71 |
| Energy contribution | -17.65 |
| Covariance contribution | 1.94 |
| Combinations/Pair | 1.23 |
| Mean z-score | -3.35 |
| Structure conservation index | 0.43 |
| SVM decision value | 2.66 |
| SVM RNA-class probability | 0.996133 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 727213 119 - 6264404 GGUGAACAGGAAGCACGCCUUUUUCCGCGCGCAAAAGCAAAACCCCCGGU-CAUCGCUGACCAGGGGUUUUGGGAUAGGAGCUUGACGAUGACCUACUCUCACAUGGGGAGACCCCACAC .((((.......((.(((........))).)).....(((((((((.(((-((....))))).)))))))))(..((((..(........).))))..)))))..(((....)))..... ( -44.10) >pau.2632 1585581 119 + 6537648 GGUGAACAGGAAGCACGCCUUUUUCCGCGCGCAAAAGCAAAACCCCUGGU-CAUCGCUGACCAGGGGUUUUGGGAUAGGAGCUUGACGAUGACCUACUCUCACAUGGGGAGACCCCACAC .((((.......((.(((........))).)).....(((((((((((((-((....)))))))))))))))(..((((..(........).))))..)))))..(((....)))..... ( -48.70) >pss.496 969900 86 - 6093698 ----------------------------------AACAAAAACCCUCAACGCGUAAGCGAUGAGGGUUUCUGGAAUUCAAUCUUGACGAUGACCUACUCUCACAUGGGGAAACCCCACAC ----------------------------------..((.(((((((((.(((....))).))))))))).))....(((.((.....))))).............(((....)))..... ( -30.10) >pfo.1282 2554707 85 - 6438405 ----------------------------------AAACAAAACCCCAACUGCUUUCGCAAUUGGGGUUUC-GGAAUUUAAUCUUGACGAUGACCUACUCUCACAUGGGGAAACCCCACAC ----------------------------------...(.(((((((((.(((....))).))))))))).-)...........(((.((........)))))...(((....)))..... ( -24.50) >consensus __________________________________AAGCAAAACCCCCAGU_CAUCGCCGACCAGGGGUUCUGGAAUAGAAGCUUGACGAUGACCUACUCUCACAUGGGGAAACCCCACAC .....................................(((((((((.(((((....))).)).)))))))))...........(((.((........)))))...(((....)))..... (-15.71 = -17.65 + 1.94)

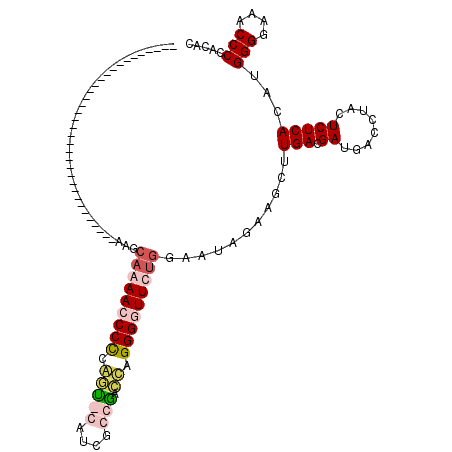
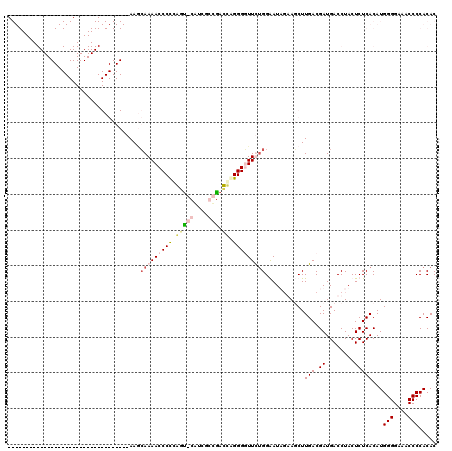
| Location | 727,228 – 727,347 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 55.52 |
| Mean single sequence MFE | -37.55 |
| Consensus MFE | -14.43 |
| Energy contribution | -15.93 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.27 |
| Mean z-score | -3.88 |
| Structure conservation index | 0.38 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980475 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 727228 119 + 6264404 AUGUGAGAGUAGGUCAUCGUCAAGCUCCUAUCCCAAAACCCCUGGUCAGCGAUG-ACCGGGGGUUUUGCUUUUGCGCGCGGAAAAAGGCGUGCUUCCUGUUCACCUUGCCGAACCCCUCC ......(((..(((..(((.((((.........(((((((((((((((....))-))))))))))))).....((((((........))))))...........)))).)))))).))). ( -47.20) >pau.2632 1585596 119 - 6537648 AUGUGAGAGUAGGUCAUCGUCAAGCUCCUAUCCCAAAACCCCUGGUCAGCGAUG-ACCAGGGGUUUUGCUUUUGCGCGCGGAAAAAGGCGUGCUUCCUGUUCACCUUGCCGAACCCCUCC ......(((..(((..(((.((((.........(((((((((((((((....))-))))))))))))).....((((((........))))))...........)))).)))))).))). ( -47.90) >pss.496 969915 71 + 6093698 AUGUGAGAGUAGGUCAUCGUCAAGAUUGAAUUCCAGAAACCCUCAUCGCUUACGCGUUGAGGGUUUUUGUU------------------------------------------------- ......((((.((((........))))..))))((((((((((((.(((....))).))))))))))))..------------------------------------------------- ( -25.80) >pfo.1282 2554722 70 + 6438405 AUGUGAGAGUAGGUCAUCGUCAAGAUUAAAUUCC-GAAACCCCAAUUGCGAAAGCAGUUGGGGUUUUGUUU------------------------------------------------- ......((((.((((........))))..))))(-((((((((((((((....)))))))))))))))...------------------------------------------------- ( -29.30) >consensus AUGUGAGAGUAGGUCAUCGUCAAGAUCCAAUCCCAAAAACCCUGAUCACCGAUG_ACCGGGGGUUUUGCUU_________________________________________________ ...(((((........)).)))...........(((((((((((((((....)).))))))))))))).....((((((........))))))........................... (-14.43 = -15.93 + 1.50)

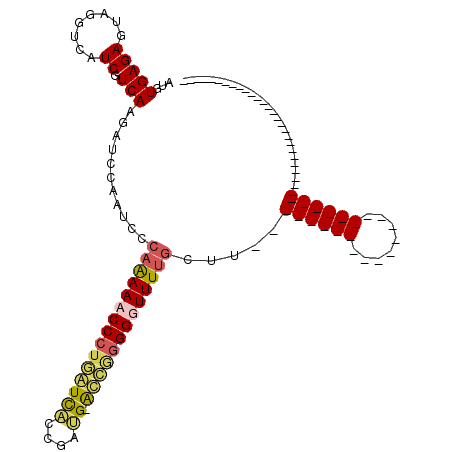
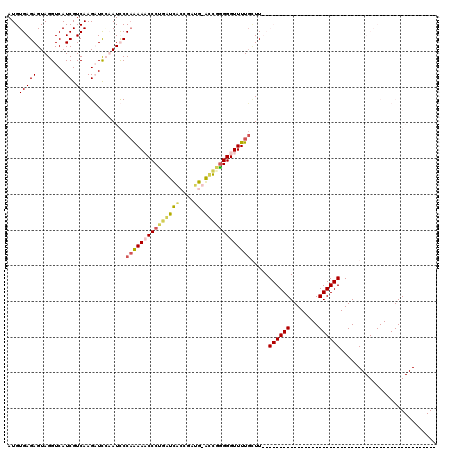
Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:08:31 2007