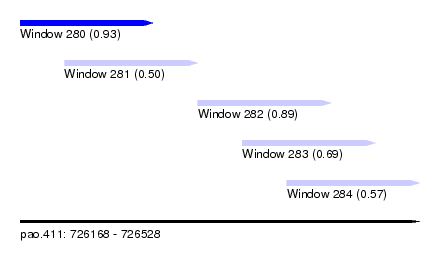
| Sequence ID | pao.411 |
|---|---|
| Location | 726,168 – 726,528 |
| Length | 360 |
| Max. P | 0.927040 |

| Location | 726,168 – 726,288 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.86 |
| Mean single sequence MFE | -43.27 |
| Consensus MFE | -43.55 |
| Energy contribution | -43.92 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.78 |
| Structure conservation index | 1.01 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.927040 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 726168 120 + 6264404 GUAGCUUUGCACUGGACUUUGAGCCUGCUUGUGUAGGAUAGGUGGGAGGCUUUGAAGCGUGGACGCCAGUUCGCGUGGAGCCAUCCUUGAAAUACCACCCUGGCAUGCUUGAGGUUCUAA .............((((((..(((.((((.(.((.((.((...(((((((((....((((((((....)))))))).))))).)))).....)))))).).)))).)))..))))))... ( -45.80) >pau.2632 1584543 120 - 6537648 GUAGCUUUGCACUGGACUUUGAGCCUGCUUGUGUAGGAUAGGUGGGAGGCUUUGAAGCGUGGACGCCAGUUCGCGUGGAGCCAUCCUUGAAAUACCACCCUGGCAUGCUUGAGGUUCUAA .............((((((..(((.((((.(.((.((.((...(((((((((....((((((((....)))))))).))))).)))).....)))))).).)))).)))..))))))... ( -45.80) >pss.496 968841 120 + 6093698 AUAGCUUUGCACUGGACUUUGAAUUUGCUUGUGUAGGAUAGGUGGGAGGCUUUGAAGCGUGGACGCCAGUCUGCGUGGAGCCAACCUUGAAAUACCACCCUGGCAACUUUGAGGUUCUAA .............((((((..((.(((((.(((((....((((....(((((....(((..(((....)))..))).))))).)))).....)).)))...))))).))..))))))... ( -40.00) >pfo.1282 2553640 120 + 6438405 AUAGCUUUGCACUGGACUUUGAAUUUGCUUGUGUAGGAUAGGUGGGAGGCUUUGAAGCGUGGACGCCAGUUCGCGUGGAGCCAUCCUUGAAAUACCACCCUGGCAACUUUGAGGUUCUAA .............((((((..((.(((((.(.((.((.((...(((((((((....((((((((....)))))))).))))).)))).....)))))).).))))).))..))))))... ( -41.50) >consensus AUAGCUUUGCACUGGACUUUGAACCUGCUUGUGUAGGAUAGGUGGGAGGCUUUGAAGCGUGGACGCCAGUUCGCGUGGAGCCAUCCUUGAAAUACCACCCUGGCAACCUUGAGGUUCUAA .............((((((..((((((((.(.((.((.((...(((((((((....((((((((....)))))))).))))).)))).....)))))).).))))))))..))))))... (-43.55 = -43.92 + 0.38)

| Location | 726,208 – 726,328 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.42 |
| Mean single sequence MFE | -46.33 |
| Consensus MFE | -42.97 |
| Energy contribution | -43.10 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.93 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>pao.411 726208 120 + 6264404 GGUGGGAGGCUUUGAAGCGUGGACGCCAGUUCGCGUGGAGCCAUCCUUGAAAUACCACCCUGGCAUGCUUGAGGUUCUAACUCUGGUCCGUAAUCCGGAUCGAGGACAGUGUAUGGUGGG ...(((((((((....((((((((....)))))))).))))).)))).......(((((...((((.((((((.......)))(((((((.....))))))))))...))))..))))). ( -47.50) >pau.2632 1584583 119 - 6537648 GGUGGGAGGCUUUGAAGCGUGGACGCCAGUUCGCGUGGAGCCAUCCUUGAAAUACCACCCUGGCAUGCUUGAGGUUCUAACUCUGGUCCGUGAUCCGGAUCGAGGACAGUGUAU-GUGGG ((((((((((((....((((((((....)))))))).))))).))))......))).(((..((((((.....(((((...(((((........)))))...)))))...))))-))))) ( -46.20) >pss.496 968881 120 + 6093698 GGUGGGAGGCUUUGAAGCGUGGACGCCAGUCUGCGUGGAGCCAACCUUGAAAUACCACCCUGGCAACUUUGAGGUUCUAACUCAGGUCCGUUAUCCGGAUCGAGGACAGUGUAUGGUGGG ...((..(((((....(((..(((....)))..))).)))))..))........(((((...(((.(((((((.......))))((((((.....))))))))).....)))..))))). ( -45.30) >pfo.1282 2553680 120 + 6438405 GGUGGGAGGCUUUGAAGCGUGGACGCCAGUUCGCGUGGAGCCAUCCUUGAAAUACCACCCUGGCAACUUUGAGGUUCUAACUCAGGUCCGUUAUCCGGAUCGAGGACAGUGUAUGGUGGG ...(((((((((....((((((((....)))))))).))))).)))).......(((((...(((.(((((((.......))))((((((.....))))))))).....)))..))))). ( -46.30) >consensus GGUGGGAGGCUUUGAAGCGUGGACGCCAGUUCGCGUGGAGCCAUCCUUGAAAUACCACCCUGGCAACCUUGAGGUUCUAACUCAGGUCCGUUAUCCGGAUCGAGGACAGUGUAUGGUGGG ...((..(((((....((((((((....)))))))).)))))..))........(((((...(((.(((((((.......))).((((((.....))))))))))....)))..))))). (-42.97 = -43.10 + 0.13)

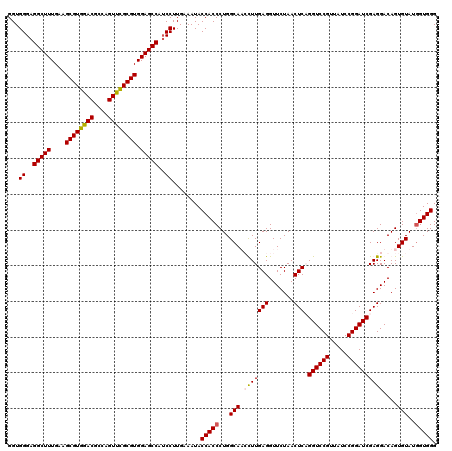
| Location | 726,328 – 726,448 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.72 |
| Mean single sequence MFE | -42.07 |
| Consensus MFE | -37.78 |
| Energy contribution | -38.65 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.891573 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 726328 120 + 6264404 CAGUUUGACUGGGGCGGUCUCCUCCUAAAGAGUAACGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGCAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGAC ..(((((.((...((((((((((((...........))))))......(((((((((..(((((((.....)))))))((...........))...))))))))))))))).)).))))) ( -44.70) >pau.2632 1584702 114 - 6537648 CAG-UUGANUGGGGCG---UCUCUCUAA--AGUAACGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGCAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGAC ...-...........(---(((((((..--......)))(.(((....(((((((((..(((((((.....)))))))((...........))...))))))))).))).)))))).... ( -35.40) >pss.496 969001 120 + 6093698 UAGUUUGACUGGGGCGGUCUCCUCCUAAAGAGUAACGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGAC ..(((((.((...((((((((((((...........))))))......(((((((((..(((((((.....))))))).....((......))...))))))))))))))).)).))))) ( -44.10) >pfo.1282 2553800 120 + 6438405 UAGUUUGACUGGGGCGGUCUCCUCCUAAAGAGUAACGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGAC ..(((((.((...((((((((((((...........))))))......(((((((((..(((((((.....))))))).....((......))...))))))))))))))).)).))))) ( -44.10) >consensus CAGUUUGACUGGGGCGGUCUCCUCCUAAAGAGUAACGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGCAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGAC ........(((..((((((((((((...........))))))......(((((((((..(((((((.....))))))).....((......))...)))))))))))))))....))).. (-37.78 = -38.65 + 0.87)

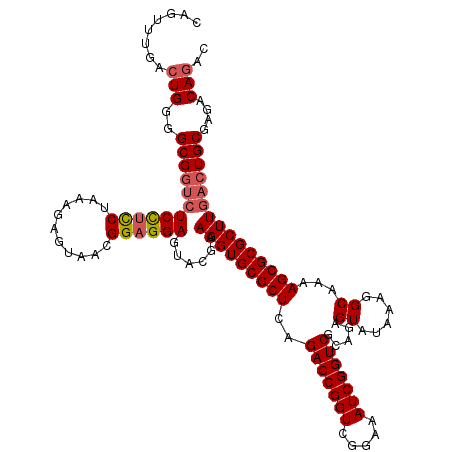
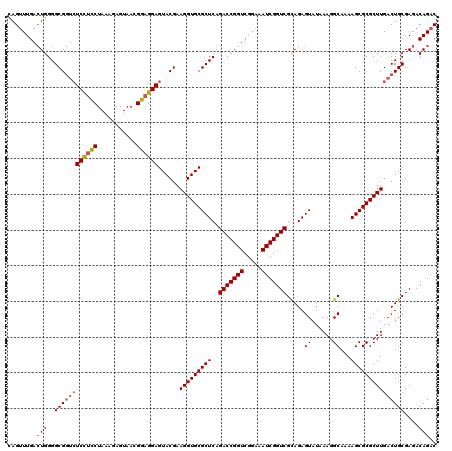
| Location | 726,368 – 726,488 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.44 |
| Mean single sequence MFE | -41.50 |
| Consensus MFE | -41.20 |
| Energy contribution | -41.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.693960 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 726368 120 + 6264404 GAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGCAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUG ..((((..(((((((((..(((((((.....)))))))((...........))...)))))))))..((((..(((......)))..)))))))).......((((.....))))..... ( -41.80) >pau.2632 1584736 120 - 6537648 GAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGCAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUG ..((((..(((((((((..(((((((.....)))))))((...........))...)))))))))..((((..(((......)))..)))))))).......((((.....))))..... ( -41.80) >pss.496 969041 120 + 6093698 GAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUG ..((((..(((((((((..(((((((.....))))))).....((......))...)))))))))..((((..(((......)))..)))))))).......((((.....))))..... ( -41.20) >pfo.1282 2553840 120 + 6438405 GAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUG ..((((..(((((((((..(((((((.....))))))).....((......))...)))))))))..((((..(((......)))..)))))))).......((((.....))))..... ( -41.20) >consensus GAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGCAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUG ..((((..(((((((((..(((((((.....))))))).....((......))...)))))))))..((((..(((......)))..)))))))).......((((.....))))..... (-41.20 = -41.20 + -0.00)

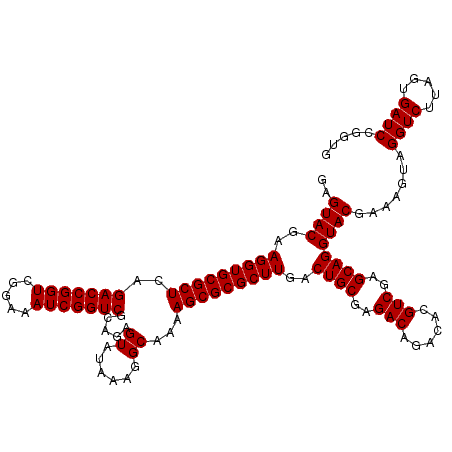
| Location | 726,408 – 726,528 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -37.00 |
| Consensus MFE | -37.00 |
| Energy contribution | -37.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.86 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.566320 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 726408 120 + 6264404 AGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGGUA .((((...(((((........((((((((((......).)).)))))))...(((....))).)))))........(((((((((.......)))))))))))))............... ( -37.00) >pau.2632 1584776 120 - 6537648 AGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGGUA .((((...(((((........((((((((((......).)).)))))))...(((....))).)))))........(((((((((.......)))))))))))))............... ( -37.00) >pss.496 969081 120 + 6093698 AGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGGUA .((((...(((((........((((((((((......).)).)))))))...(((....))).)))))........(((((((((.......)))))))))))))............... ( -37.00) >pfo.1282 2553880 120 + 6438405 AGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGGUA .((((...(((((........((((((((((......).)).)))))))...(((....))).)))))........(((((((((.......)))))))))))))............... ( -37.00) >consensus AGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGGUA .((((...(((((........((((((((((......).)).)))))))...(((....))).)))))........(((((((((.......)))))))))))))............... (-37.00 = -37.00 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:08:23 2007