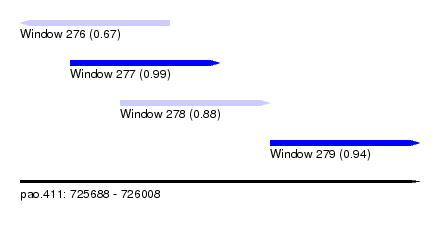
| Sequence ID | pao.411 |
|---|---|
| Location | 725,688 – 726,008 |
| Length | 320 |
| Max. P | 0.994402 |

| Location | 725,688 – 725,808 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.50 |
| Mean single sequence MFE | -36.35 |
| Consensus MFE | -35.20 |
| Energy contribution | -35.45 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.670060 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 725688 120 - 6264404 CCUAGCCGGCGCACCUUCUCCCGAAGUUACGGUGCCAUUUUGCCUAGUUCCUUCACCCGAGUUCUCUCAAGCGCCUUGGUAUUCUCUACCCGACCACCUGUGUCGGUUUGGGGUACGGUU ...(((((((((..........((((..(((((........)))..))..))))....(((....)))..)))))...(((((((..(((.((((....).))))))..))))))))))) ( -38.40) >pau.2632 1584063 120 + 6537648 CCUAGCCGGCGCACCUUCUCCCGAAGUUACGGUGCCAUUUUGCCUAGUUCCUUCACCCGAGUUCUCUCAAGCGCCUUGGUAUUCUCUACCCGACCACCUGUGUCGGUUUGGGGUACGGUU ...(((((((((..........((((..(((((........)))..))..))))....(((....)))..)))))...(((((((..(((.((((....).))))))..))))))))))) ( -38.40) >pss.496 968361 120 - 6093698 CUCCACCGGCGCACCUUCUCCCGAAGUUACGGUGCCAUUUUGCCUAGUUCCUUCACCCGAGUUCUCUCAAGCGCCUUGGUAUUCUCUACCCAACCACCUGUGUCGGUUUGGGGUACGGUU ....((((((((..........((((..(((((........)))..))..))))....(((....)))..)))))...(((((((..(((..((.....))...)))..)))))))))). ( -34.30) >pfo.1282 2553160 120 - 6438405 CCUCACCGGCGCACCUUCUCCCGAAGUUACGGUGCCAUUUUGCCUAGUUCCUUCACCCGAGUUCUCUCAAGCGCCUUGGUAUUCUCUACCCAACCACCUGUGUCGGUUUGGGGUACGGUU ....((((((((..........((((..(((((........)))..))..))))....(((....)))..)))))...(((((((..(((..((.....))...)))..)))))))))). ( -34.30) >consensus CCUAACCGGCGCACCUUCUCCCGAAGUUACGGUGCCAUUUUGCCUAGUUCCUUCACCCGAGUUCUCUCAAGCGCCUUGGUAUUCUCUACCCAACCACCUGUGUCGGUUUGGGGUACGGUU ....((((((((..........((((..(((((........)))..))..))))....(((....)))..)))))...(((((((..(((.((((....).))))))..)))))))))). (-35.20 = -35.45 + 0.25)

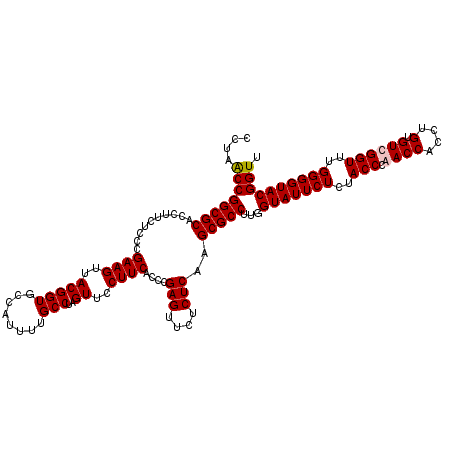
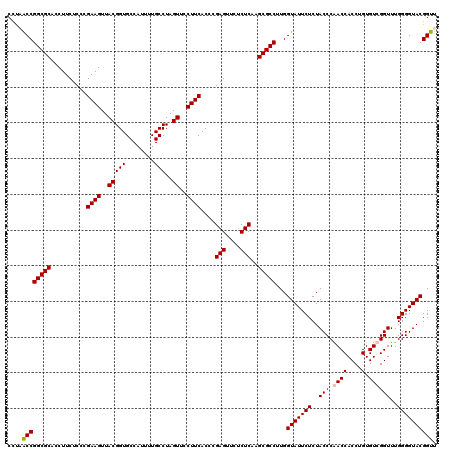
| Location | 725,728 – 725,848 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.61 |
| Mean single sequence MFE | -44.45 |
| Consensus MFE | -44.56 |
| Energy contribution | -43.00 |
| Covariance contribution | -1.56 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.40 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.48 |
| SVM RNA-class probability | 0.994402 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 725728 120 + 6264404 ACCAAGGCGCUUGAGAGAACUCGGGUGAAGGAACUAGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGCUAGGGUGAAGGAUUUACUCCGUAAGCUCUGGCUGGUCGAAGAUA .((....((((((((....))))))))..))..............(((((....(....).....)))))((((((((((((...(((.....)))....))))))))))))........ ( -45.20) >pau.2632 1584103 120 - 6537648 ACCAAGGCGCUUGAGAGAACUCGGGUGAAGGAACUAGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGCUAGGGUGAAGGAUUUACUCCGUAAGCUCUGGCUGGUCGAAGAUA .((....((((((((....))))))))..))..............(((((....(....).....)))))((((((((((((...(((.....)))....))))))))))))........ ( -45.20) >pss.496 968401 120 + 6093698 ACCAAGGCGCUUGAGAGAACUCGGGUGAAGGAACUAGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGUGGAGGUGAAGCAUUUACUGCGUAAGCCCCUGCCGGUCGAAGAUA .((....((((((((....))))))))..))..............(((((....(....).....)))))(((((..(.(((...(((.....)))....))).)..)))))........ ( -42.40) >pfo.1282 2553200 120 + 6438405 ACCAAGGCGCUUGAGAGAACUCGGGUGAAGGAACUAGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGUGAGGGUGAAGCACUUGCUGCGUAAGCCCACGCCGGUCGAAGAUA .((....((((((((....))))))))..))..............(((((....(....).....)))))(((((((.((((...(((.....)))....)))).)))))))........ ( -45.00) >consensus ACCAAGGCGCUUGAGAGAACUCGGGUGAAGGAACUAGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGCGAGGGUGAAGCAUUUACUCCGUAAGCCCUGGCCGGUCGAAGAUA .((....((((((((....))))))))..))..............(((((.....(((....))))))))((((((((((((...(((.....)))....))))))))))))........ (-44.56 = -43.00 + -1.56)

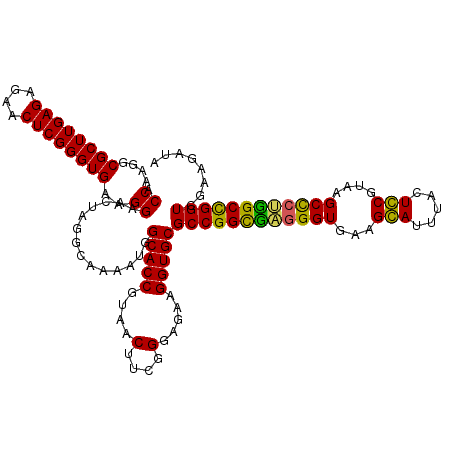
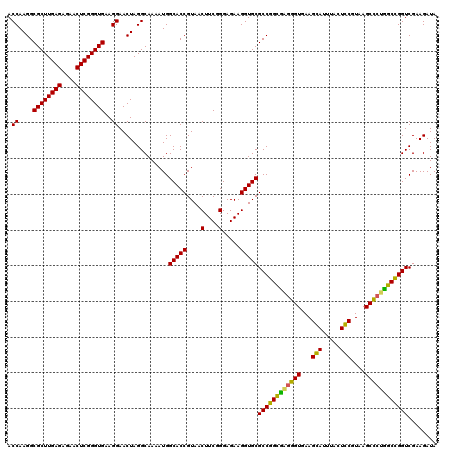
| Location | 725,768 – 725,888 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.61 |
| Mean single sequence MFE | -42.25 |
| Consensus MFE | -43.96 |
| Energy contribution | -42.40 |
| Covariance contribution | -1.56 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.54 |
| Structure conservation index | 1.04 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.879882 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 725768 120 + 6264404 AAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGCUAGGGUGAAGGAUUUACUCCGUAAGCUCUGGCUGGUCGAAGAUACCAGGCCGCUGCGACUGUUUAUUAAAAACACAGCACUCUG ....(((...(((.((((....)))).)))((((((((((((...(((.....)))....))))))))))))............)))((((....(((((.....)))))))))...... ( -43.00) >pau.2632 1584143 120 - 6537648 AAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGCUAGGGUGAAGGAUUUACUCCGUAAGCUCUGGCUGGUCGAAGAUACCAGGCCGCUGCGACUGUUUAUUAAAAACACAGCACUCUG ....(((...(((.((((....)))).)))((((((((((((...(((.....)))....))))))))))))............)))((((....(((((.....)))))))))...... ( -43.00) >pss.496 968441 120 + 6093698 AAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGUGGAGGUGAAGCAUUUACUGCGUAAGCCCCUGCCGGUCGAAGAUACCAGGCCGCUGCGACUGUUUAUUAAAAACACAGCACUCUG ....(((...(((.((((....)))).)))(((((..(.(((...(((.....)))....))).)..)))))............)))((((....(((((.....)))))))))...... ( -40.20) >pfo.1282 2553240 120 + 6438405 AAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGUGAGGGUGAAGCACUUGCUGCGUAAGCCCACGCCGGUCGAAGAUACCAGGCCGCUGCGACUGUUUAUUAAAAACACAGCACUCUG ....(((...(((.((((....)))).)))(((((((.((((...(((.....)))....)))).)))))))............)))((((....(((((.....)))))))))...... ( -42.80) >consensus AAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGCGAGGGUGAAGCAUUUACUCCGUAAGCCCUGGCCGGUCGAAGAUACCAGGCCGCUGCGACUGUUUAUUAAAAACACAGCACUCUG ....(((...(((.((((....)))).)))((((((((((((...(((.....)))....))))))))))))............)))((((....(((((.....)))))))))...... (-43.96 = -42.40 + -1.56)

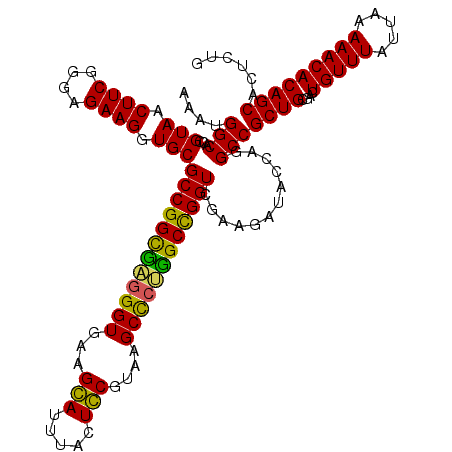
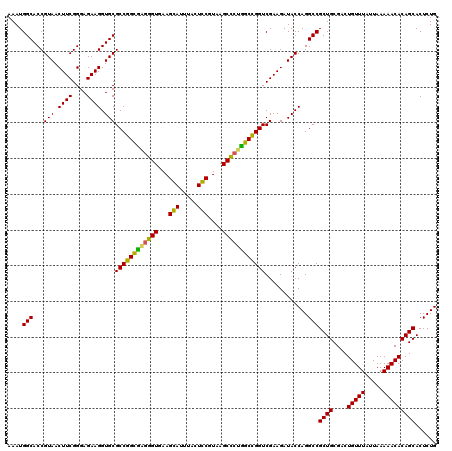
| Location | 725,888 – 726,008 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.75 |
| Mean single sequence MFE | -46.25 |
| Consensus MFE | -45.75 |
| Energy contribution | -46.25 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940499 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.411 725888 120 + 6264404 CAAACACGAAAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCGCAAGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGGCGGCCGUAACUAUAA ....(((....)))....(((((......(((.(((((...((((((..((((..(((((((((((.((....))..))))))..)))))))))))))))...))))).)))..))))). ( -46.70) >pau.2632 1584263 120 - 6537648 CAAACACGAAAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCGCAAGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGGCGGCCGUAACUAUAA ....(((....)))....(((((......(((.(((((...((((((..((((..(((((((((((.((....))..))))))..)))))))))))))))...))))).)))..))))). ( -46.70) >pss.496 968561 120 + 6093698 CAAACACGAAAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCGCAAGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGGCGGCCGUAACUAUAA ....(((....)))....(((((......(((.(((((...((((((..((((..(((((((((((.((....))..))))))..)))))))))))))))...))))).)))..))))). ( -46.70) >pfo.1282 2553360 120 + 6438405 CAAACACGAAAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCUAACGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGGCGGCCGUAACUAUAA ....(((....)))....(((((......(((.(((((...((((((..((((..(((((((((((.((....))..))))))..)))))))))))))))...))))).)))..))))). ( -44.90) >consensus CAAACACGAAAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCGCAAGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGGCGGCCGUAACUAUAA ....(((....)))....(((((......(((.(((((...((((((..((((..(((((((((((.((....))..))))))..)))))))))))))))...))))).)))..))))). (-45.75 = -46.25 + 0.50)


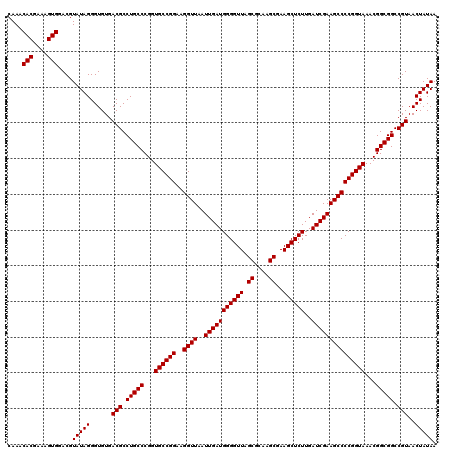
Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:08:17 2007